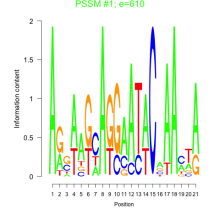
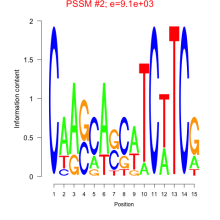
Thaps_bicluster_0218 Residual: 0.15
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0218 | 0.15 | Thalassiosira pseudonana |
Displaying 1 - 14 of 14
" class="views-fluidgrid-wrapper clear-block">
20603 hypothetical protein
24099 hypothetical protein
24769 hypothetical protein
2601 (Lhca2) Chloroa_b-bind
268127 (Lhcf9) PLN00120
268304 hypothetical protein
2848 PsbU
29842 Rieske_2
30385 (Lhcx6_1) Chloroa_b-bind superfamily
3141 hypothetical protein
34830 MSP superfamily
38583 (Lhcf1) PLN00120
42962 (Lhcf5) PLN00120
5174 (Lhcf8) PLN00120
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009765 | photosynthesis, light harvesting | 0.00454357 | 1 | Thaps_bicluster_0218 |
| GO:0042549 | photosystem II stabilization | 0.0000281 | 0.0774835 | Thaps_bicluster_0218 |
| GO:0015979 | photosynthesis | 0.0291995 | 1 | Thaps_bicluster_0218 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0218 |

Comments