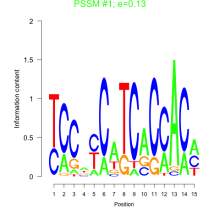
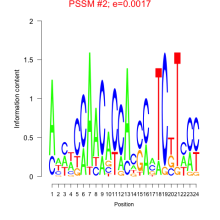
Thaps_bicluster_0226 Residual: 0.29
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0226 | 0.29 | Thalassiosira pseudonana |
Displaying 1 - 19 of 19
" class="views-fluidgrid-wrapper clear-block">
20737 Sun
2157 D123 superfamily
21973 G-patch
22861 hypothetical protein
22999 hypothetical protein
23209 hypothetical protein
24439 hypothetical protein
24907 Sas10
25183 hypothetical protein
262325 SpoU_methylase
268880 WD40 superfamily
268970 PGDH_like_3
269975 (Tp_TAZ2) regulator [Rayko]
28300 dimethyladenosine transferase
3018 AAT_I
32790 RRM_eIF3G_like
5104 hypothetical protein
6020 DUF2088 superfamily
9118 MoaC_PE
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0000154 | rRNA modification | 0.00289256 | 1 | Thaps_bicluster_0226 |
| GO:0006777 | Mo-molybdopterin cofactor biosynthesis | 0.00865617 | 1 | Thaps_bicluster_0226 |
| GO:0006364 | rRNA processing | 0.0115273 | 1 | Thaps_bicluster_0226 |
| GO:0006564 | L-serine biosynthesis | 0.0129601 | 1 | Thaps_bicluster_0226 |
| GO:0009058 | biosynthesis | 0.0769767 | 1 | Thaps_bicluster_0226 |
| GO:0006396 | RNA processing | 0.081025 | 1 | Thaps_bicluster_0226 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.368277 | 1 | Thaps_bicluster_0226 |

Comments