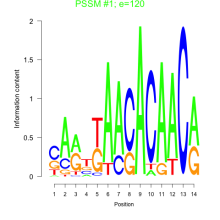
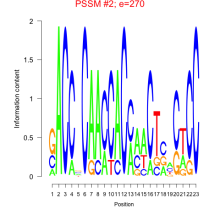
Thaps_bicluster_0229 Residual: 0.40
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0229 | 0.40 | Thalassiosira pseudonana |
Displaying 1 - 35 of 35
" class="views-fluidgrid-wrapper clear-block">
10129 Dtyr_deacylase superfamily
11342 RNA_pol_Rpc4 superfamily
13982 SrmB
1692 hypothetical protein
18538 Dynein_heavy superfamily
20962 hypothetical protein
20972 hypothetical protein
21281 hypothetical protein
21800 Peptidase_M14_like superfamily
22385 SAC3_GANP
22623 KH-I
22978 Aar2_C
23810 (PPS1) PRK06464
24017 hypothetical protein
2443 hypothetical protein
24820 hypothetical protein
25725 TatA
25906 hypothetical protein
260906 pyridoxal_pyridoxamine_kinase
261260 PHO4
261802 Aldo_ket_red
268574 Tim17
268661 (pab1) RRM_SF superfamily
268872 SET
269976 SRPBCC superfamily
2941 hypothetical protein
30550 Utp11
31783 PrmA
34730 carb_red_sniffer_like_SDR_c
35274 RRM_Srp1p_AtRSp31_like
35748 Dynein_heavy superfamily
36958 PKc_like superfamily
5497 hypothetical protein
6648 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019478 | D-amino acid catabolism | 0.00351458 | 1 | Thaps_bicluster_0229 |
| GO:0006399 | tRNA metabolism | 0.00351458 | 1 | Thaps_bicluster_0229 |
| GO:0015031 | protein transport | 0.00964735 | 1 | Thaps_bicluster_0229 |
| GO:0006817 | phosphate transport | 0.017457 | 1 | Thaps_bicluster_0229 |
| GO:0006364 | rRNA processing | 0.027793 | 1 | Thaps_bicluster_0229 |
| GO:0006094 | gluconeogenesis | 0.0312156 | 1 | Thaps_bicluster_0229 |
| GO:0016310 | phosphorylation | 0.0447927 | 1 | Thaps_bicluster_0229 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0779563 | 1 | Thaps_bicluster_0229 |
| GO:0006350 | transcription | 0.100523 | 1 | Thaps_bicluster_0229 |
| GO:0008152 | metabolism | 0.167146 | 1 | Thaps_bicluster_0229 |

Comments