Thaps_bicluster_0269 Residual: 0.41
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0269 | 0.41 | Thalassiosira pseudonana |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
10624 hypothetical protein
10810 HP superfamily
21239 Cyt_b561_FRRS1_like
21948 DUF179
22349 hypothetical protein
23170 hypothetical protein
23618 RING
261381 Rab5_related
261827 PDI_a_family
263742 Synaptobrevin
27352 PLN00158
27377 Peptidase_S9
32945 (SDH1) sdhB
3359 hypothetical protein
3426 zf-DNL superfamily
34738 AAA_12 superfamily
35407 PLN00157
36076 HDAC_classIV
36764 PRK05279
39847 (APM2) AP-2_Mu2_Cterm
40586 (GMD1) PLN02653
4774 hypothetical protein
7012 hypothetical protein
8058 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 0.00616834 | 1 | Thaps_bicluster_0269 |
| GO:0006334 | nucleosome assembly | 0.00702986 | 1 | Thaps_bicluster_0269 |
| GO:0006886 | intracellular protein transport | 0.0204396 | 1 | Thaps_bicluster_0269 |
| GO:0006526 | arginine biosynthesis | 0.0366334 | 1 | Thaps_bicluster_0269 |
| GO:0006099 | tricarboxylic acid cycle | 0.0406241 | 1 | Thaps_bicluster_0269 |
| GO:0016192 | vesicle-mediated transport | 0.0642408 | 1 | Thaps_bicluster_0269 |
| GO:0007264 | small GTPase mediated signal transduction | 0.131822 | 1 | Thaps_bicluster_0269 |
| GO:0006096 | glycolysis | 0.142624 | 1 | Thaps_bicluster_0269 |
| GO:0015031 | protein transport | 0.163848 | 1 | Thaps_bicluster_0269 |
| GO:0009058 | biosynthesis | 0.204806 | 1 | Thaps_bicluster_0269 |

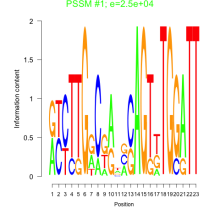

Comments