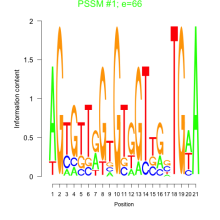
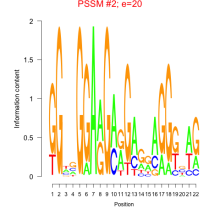
Thaps_bicluster_0278 Residual: 0.24
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0278 | 0.24 | Thalassiosira pseudonana |
Displaying 1 - 13 of 13
" class="views-fluidgrid-wrapper clear-block">
11949 Nop14
18557 HrpA
21368 hypothetical protein
21375 hypothetical protein
21805 hypothetical protein
23079 zf-LYAR
25436 hypothetical protein
268746 SXM1
34661 COG4581
34881 Pescadillo_N superfamily
36798 S_TKc
40529 COG2319
8336 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008283 | cell proliferation | 0.00206654 | 1 | Thaps_bicluster_0278 |
| GO:0000059 | protein-nucleus import, docking | 0.00515995 | 1 | Thaps_bicluster_0278 |
| GO:0006397 | mRNA processing | 0.0225447 | 1 | Thaps_bicluster_0278 |
| GO:0006468 | protein amino acid phosphorylation | 0.201698 | 1 | Thaps_bicluster_0278 |
| GO:0006508 | proteolysis and peptidolysis | 0.264399 | 1 | Thaps_bicluster_0278 |

Comments