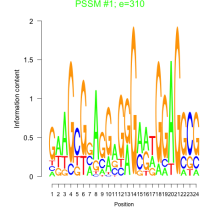
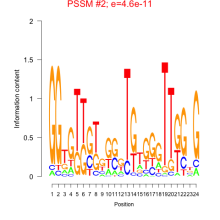
Thaps_bicluster_0287 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0287 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
11325 Aa_trans superfamily
1383 (PDS1) phytoene_desat
13922 2A0303
16605 Aldolase_II superfamily
1773 hypothetical protein
20921 S2P-M50_like_2
21360 hypothetical protein
225 RRF
261690 ABCG_EPDR
262510 Alg6_Alg8 superfamily
262572 Octopine_DH
262762 BET4
263431 Ptr
263724 sigpep_I_bact
263781 sulP
3577 hypothetical protein
36494 NBD_sugar-kinase_HSP70_actin superfamily
38240 HA
38312 PTZ00318
5286 Cupin_8 superfamily
6056 GCD2
6897 RhaT
7191 Aldo_ket_red
9000 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006865 | amino acid transport | 0.00184627 | 1 | Thaps_bicluster_0287 |
| GO:0018346 | protein amino acid prenylation | 0.0113237 | 1 | Thaps_bicluster_0287 |
| GO:0006508 | proteolysis and peptidolysis | 0.0241111 | 1 | Thaps_bicluster_0287 |
| GO:0006413 | translational initiation | 0.053299 | 1 | Thaps_bicluster_0287 |
| GO:0006118 | electron transport | 0.119465 | 1 | Thaps_bicluster_0287 |
| GO:0006412 | protein biosynthesis | 0.319237 | 1 | Thaps_bicluster_0287 |
| GO:0006810 | transport | 0.347574 | 1 | Thaps_bicluster_0287 |

Comments