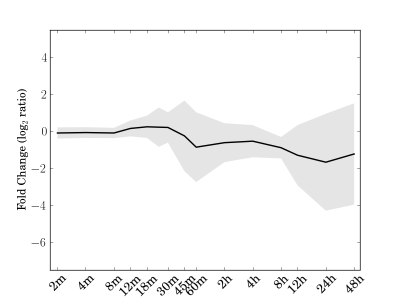
Module 13 Residual: 0.15
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0013 | v02 | 0.15 | -12.28 |
Displaying 1 - 26 of 26
| Cre01.g004124.t2.1 | Cre01.g007901.t1.1 MATE efflux family protein |
| Cre01.g016850.t1.2 Uncharacterised protein family SERF | Cre02.g073650.t2.1 MOS4-associated complex 3B |
| Cre02.g107200.t1.1 | Cre03.g174350.t1.2 Protein of unknown function (DUF506) |
| Cre04.g223100.t1.2 alpha carbonic anhydrase 4 | Cre06.g278239.t1.1 SERINE-ARGININE PROTEIN 30 |
| Cre07.g342551.t1.1 ubiquitin-conjugating enzyme 30 | Cre07.g348550.t2.1 alpha/beta-Hydrolases superfamily protein |
| Cre08.g374400.t2.1 Transducin/WD40 repeat-like superfamily protein | Cre09.g398500.t1.1 |
| Cre10.g422800.t1.1 | Cre10.g427200.t1.1 |
| Cre10.g429000.t2.1 | Cre10.g452350.t1.1 |
| Cre12.g530850.t2.1 | Cre14.g615250.t1.1 Thioredoxin superfamily protein |
| Cre14.g617400.t1.2 HSP20-like chaperones superfamily protein | Cre14.g620200.t1.1 |
| Cre16.g654250.t1.2 | Cre16.g676350.t1.1 Cam interacting protein 111 |
| Cre16.g680700.t1.2 Aluminium induced protein with YGL and LRDR motifs | Cre16.g690431.t2.1 |
| Cre17.g725750.t1.2 | Cre17.g746597.t2.1 serine carboxypeptidase-like 20 |
|
e.value: 0.0000011 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: TgCTGCTGctGCTGCtGCTGc |
motif_0013_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.045 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: gTGcagGTGtAGgTG |
Displaying 1 - 3 of 3
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre10.g453500.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.067406 |
| up-regulates |
Cre03.g152150.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.038235 |
| up-regulates |
Cre12.g534450.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51SMAD/FHA domain-containing protein |
0.038806 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

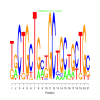
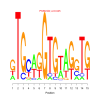
Comments