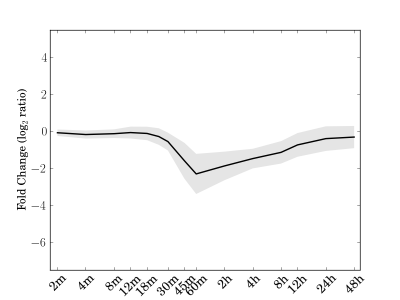
Module 40 Residual: 0.25
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0040 | v02 | 0.25 | -12.73 |
Displaying 1 - 25 of 25
| Cre01.g006100.t1.2 | Cre01.g014050.t1.2 Zinc finger C-x8-C-x5-C-x3-H type family protein |
| Cre01.g017500.t1.2 sequence-specific DNA binding transcription factors | Cre01.g039350.t1.2 P450 reductase 1 |
| Cre01.g055453.t1.1 VALINE-TOLERANT 1 | Cre02.g086550.t1.1 Radical SAM superfamily protein |
| Cre02.g103850.t1.2 HISTIDINE BIOSYNTHESIS 5B | Cre03.g177200.t1.2 RNA-binding (RRM/RBD/RNP motifs) family protein |
| Cre03.g192050.t1.1 Iron permease, membrane protein | Cre03.g194400.t1.2 Transducin/WD40 repeat-like superfamily protein |
| Cre03.g195000.t1.2 | Cre05.g238250.t1.2 Basic-leucine zipper (bZIP) transcription factor family protein |
| Cre06.g301650.t1.2 | Cre06.g310200.t1.2 tetratricopeptide repeat (TPR)-containing protein |
| Cre08.g372850.t1.2 ZPR1 zinc-finger domain protein | Cre09.g403500.t1.2 XB3 ortholog 1 in Arabidopsis thaliana |
| Cre10.g448800.t1.1 Ribosomal protein S5 domain 2-like superfamily protein | Cre11.g467633.t1.1 Methylthiotransferase |
| Cre13.g569250.t1.2 Predicted protein | Cre13.g588150.t1.2 galactose-1-phosphate guanylyltransferase (GDP)s;GDP-D-glucose phosphorylases;quercetin 4\'-O-glucosyltransferases |
| Cre13.g588700.t1.1 | Cre14.g615950.t1.1 general control non-repressible 4 |
| Cre16.g683550.t1.2 P450 reductase 1 | Cre16.g688050.t1.1 eukaryotic translation initiation factor 4B1 |
| Cre17.g734564.t1.1 |
|
e.value: 0.000073 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: aCaGcaaCAgcaGCAgcgGCAGca |
motif_0040_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.77 Motif Bicluster: Width: 23 Number of Sites: 1 Consensus: GTTTgCTGaTGCgCATTcCtCAt |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre13.g573000.t1.2Submitted by admin on Tue, 05/19/2015 - 15:52SET domain-containing protein |
0.054006 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0010181 | FMN binding |


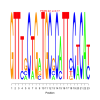
Comments