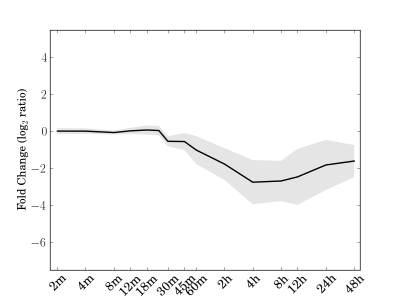
Module 58 Residual: 0.17
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0058 | v02 | 0.17 | -9.70 |
Displaying 1 - 19 of 19
| Cre01.g000850.t1.2 Predicted protein | Cre01.g004300.t1.2 glutamine-dependent asparagine synthase 1 |
| Cre01.g069472.t1.1 Cysteinyl-tRNA synthetase, class Ia family protein | Cre02.g076300.t1.1 Uroporphyrinogen decarboxylase |
| Cre02.g085450.t1.2 Coproporphyrinogen III oxidase | Cre02.g092400.t1.1 Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain |
| Cre02.g100050.t1.2 Oxoglutarate/iron-dependent oxygenase | Cre02.g109550.t1.2 aspartate kinase-homoserine dehydrogenase ii |
| Cre02.g114400.t1.2 Radical SAM superfamily protein | Cre03.g146167.t1.1 Predicted protein |
| Cre06.g300700.t1.1 NAD(P)-binding Rossmann-fold superfamily protein | Cre07.g325734.t1.1 nucleoside diphosphate kinase 2 |
| Cre09.g393173.t1.1 Chlorophyll A-B binding family protein | Cre10.g430150.t1.2 tetratricopeptide repeat (TPR)-containing protein |
| Cre10.g457650.t1.2 secE/sec61-gamma protein transport protein | Cre10.g460050.t2.1 Tetratricopeptide repeat (TPR)-like superfamily protein |
| Cre12.g519100.t1.2 acetyl Co-enzyme a carboxylase carboxyltransferase alpha subunit | Cre13.g576760.t1.1 Chlorophyll A-B binding family protein |
| Cre13.g578650.t1.1 high chlorophyll fluorescence phenotype 173 |
|
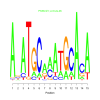
e.value: 0.0000092 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: AnATGCaaATGCAcA |
motif_0058_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 36 Motif Bicluster: Width: 8 Number of Sites: 1 Consensus: CAAcCAAA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006082 | GO:0019752 | GO:0043436 | GO:0009317 | organic acid metabolic process | carboxylic acid metabolic process | oxoacid metabolic process | acetyl-CoA carboxylase complex |


Comments