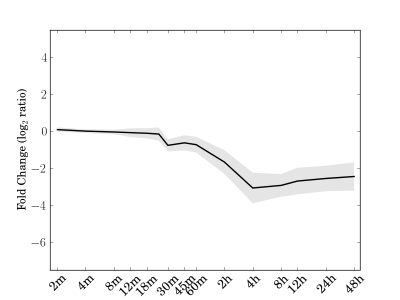
Module 69 Residual: 0.14
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0069 | v02 | 0.14 | -14.92 |
Displaying 1 - 16 of 16
| Cre01.g016514.t1.1 lipoamide dehydrogenase 1 | Cre01.g040300.t1.2 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| Cre03.g156600.t1.2 proton gradient regulation 7 | Cre03.g158000.t1.2 glutamate-1-semialdehyde 2,1-aminomutase 2 |
| Cre03.g199535.t1.1 Chlorophyll A-B binding family protein | Cre04.g213251.t1.1 Peptidase M1 family protein |
| Cre05.g242000.t1.2 ALBINA 1 | Cre05.g246800.t1.2 enzyme binding;tetrapyrrole binding |
| Cre06.g306300.t1.2 P-loop containing nucleoside triphosphate hydrolases superfamily protein | Cre07.g328200.t1.2 Mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein |
| Cre12.g490500.t1.2 Predicted protein | Cre12.g510800.t1.2 P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| Cre12.g519300.t1.2 Predicted protein | Cre12.g551950.t1.2 Ankyrin repeat family protein |
| Cre16.g663900.t1.2 hydroxymethylbilane synthase | Cre17.g702200.t1.2 ankyrin repeat-containing protein 2 |
|
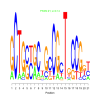
e.value: 0.14 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: GCTGctgCtGcCGcTgCCGnt |
motif_0069_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 8.5 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: aATGaAAcagTGcaA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0016874 | ligase activity |

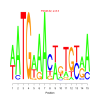
Comments