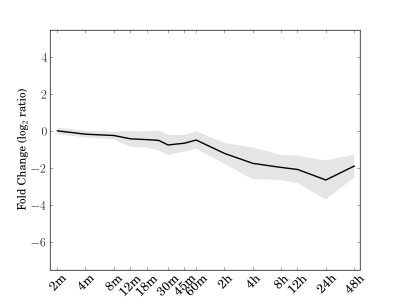
Module 78 Residual: 0.29
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0078 | v02 | 0.29 | -15.59 |
Displaying 1 - 20 of 20
| Cre01.g005950.t1.2 FAD/NAD(P)-binding oxidoreductase | Cre01.g013350.t1.2 |
| Cre01.g014250.t1.1 | Cre01.g015350.t1.1 protochlorophyllide oxidoreductase A |
| Cre01.g022500.t1.2 NADP-malic enzyme 4 | Cre01.g024200.t1.1 cleavage stimulating factor 64 |
| Cre04.g213761.t2.1 Thioesterase superfamily protein | Cre04.g219787.t1.2 UbiA prenyltransferase family protein |
| Cre05.g243050.t1.2 thioredoxin F-type 1 | Cre06.g255350.t1.2 hydroxyethylthiazole kinase family protein |
| Cre07.g321300.t1.2 NAD(P)-linked oxidoreductase superfamily protein | Cre08.g377100.t1.2 adenosine kinase |
| Cre09.g396250.t1.2 vitamin E pathway gene 5 | Cre12.g519050.t1.1 |
| Cre12.g543900.t1.1 | Cre14.g626500.t1.2 Transducin/WD40 repeat-like superfamily protein |
| Cre16.g672350.t1.1 breast cancer susceptibility1 | Cre16.g674300.t1.1 ankyrin repeat-containing protein 2 |
| Cre17.g700750.t1.2 | Cre17.g714900.t1.2 thylakoid lumenal 17.9 kDa protein, chloroplast |
|
e.value: 0.000000056 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: GTgtGcgTGtgcgTGgGTGtG |
motif_0078_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000000000000017 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: gCtcCtGCtgCaGCTcctgCagCt |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |



Comments