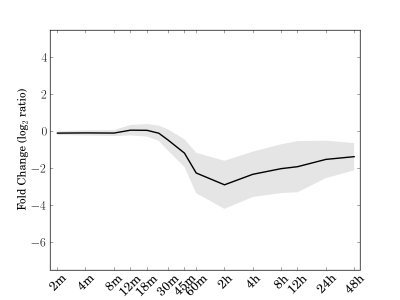
Module 105 Residual: 0.21
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0105 | v02 | 0.21 | -13.29 |
Displaying 1 - 22 of 22
| Cre01.g016850.t1.2 Uncharacterised protein family SERF | Cre01.g039350.t1.2 P450 reductase 1 |
| Cre02.g077350.t1.2 histidinol dehydrogenase | Cre02.g078100.t1.1 eukaryotic translation initiation factor 2 (eIF-2) family protein |
| Cre02.g081050.t1.2 Flagellar Associated Protein | Cre03.g189400.t1.2 seryl-tRNA synthetase / serine--tRNA ligase |
| Cre03.g206600.t1.2 dehydratase family | Cre04.g231222.t2.1 chaperonin-60alpha |
| Cre05.g245900.t1.2 branched-chain amino acid aminotransferase 5 / branched-chain amino acid transaminase 5 (BCAT5) | Cre06.g250902.t1.1 Homocysteine S-methyltransferase family protein |
| Cre06.g299000.t1.2 Ribosomal protein L21 | Cre10.g436350.t1.1 shikimate kinase 1 |
| Cre10.g439900.t1.2 heat shock protein 70 (Hsp 70) family protein | Cre11.g479150.t3.1 |
| Cre12.g494750.t1.2 chloroplast 30S ribosomal protein S20, putative | Cre12.g501550.t1.2 |
| Cre13.g603900.t1.2 tRNA synthetase beta subunit family protein | Cre14.g611450.t1.1 Plastid-lipid associated protein PAP / fibrillin family protein |
| Cre16.g651750.t1.2 tRNA synthetase class I (I, L, M and V) family protein | Cre16.g693850.t1.1 |
| Cre17.g697406.t1.2 | Cre17.g734450.t1.2 Ribosomal protein L19 family protein |
|
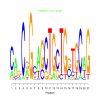
e.value: 0.000000014 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CagCaGcAGCTGcTGgTGCaG |
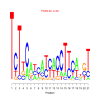
motif_0105_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 82 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: TcTtcatcatcacCTtcangT |
Displaying 1 - 2 of 2
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre12.g501600.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51Basic-leucine zipper (bZIP) transcription factor family protein |
0.054978 |
| up-regulates |
Cre17.g746547.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51Basic-leucine zipper (bZIP) transcription factor family protein |
0.05663 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006520 | GO:0006082 | GO:0019752 | GO:0043436 | GO:0006399 | GO:0034645 | GO:0009059 | GO:0034660 | GO:0005737 | cellular amino acid metabolic process | organic acid metabolic process | carboxylic acid metabolic process | oxoacid metabolic process | tRNA metabolic process | cellular macromolecule biosynthetic proc... | macromolecule biosynthetic process | ncRNA metabolic process | cytoplasm |

Comments