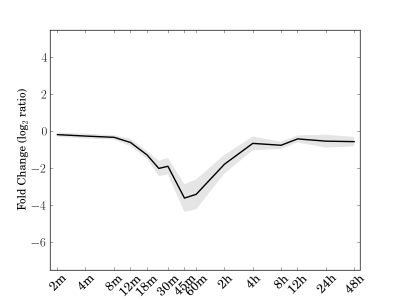
Module 108 Residual: 0.12
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0108 | v02 | 0.12 | -18.31 |
Displaying 1 - 17 of 17
| Cre02.g092100.t1.2 | Cre02.g093950.t1.2 Amino acid kinase family protein |
| Cre02.g117900.t1.1 RNA helicase, ATP-dependent, SK12/DOB1 protein | Cre04.g217933.t1.1 |
| Cre06.g278175.t1.1 Transducin/WD40 repeat-like superfamily protein | Cre07.g314900.t1.1 P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| Cre09.g388500.t1.2 | Cre10.g430450.t1.2 formyltetrahydrofolate deformylase, putative |
| Cre10.g454100.t1.2 Ribosomal RNA processing Brix domain protein | Cre11.g480060.t1.2 Polynucleotidyl transferase, ribonuclease H-like superfamily protein |
| Cre12.g499000.t1.2 | Cre12.g505200.t1.2 DEA(D/H)-box RNA helicase family protein |
| Cre12.g508200.t1.2 RNA-binding KH domain-containing protein | Cre12.g512550.t1.2 Trm112p-like protein |
| Cre16.g649400.t1.2 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Cre16.g649600.t1.2 pumilio 23 |
| Cre17.g700650.t1.2 |
|
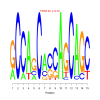
e.value: 0.000000000000068 Motif Bicluster: Width: 23 Number of Sites: 1 Consensus: CTGCtGCcgcTGCTgCTGCtgCa |
motif_0108_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.02 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: GCCAgCacCAGCAGC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |


Comments