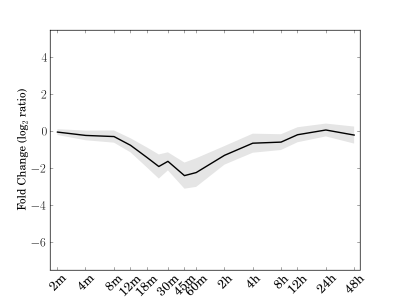
Module 123 Residual: 0.24
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0123 | v02 | 0.24 | -12.46 |
Displaying 1 - 24 of 24
| Cre01.g019200.t1.1 | Cre01.g024100.t1.1 |
| Cre01.g036700.t1.2 NPK1-related protein kinase 2 | Cre01.g043000.t1.1 Radical SAM superfamily protein |
| Cre01.g049600.t1.2 ATP synthase protein I -related | Cre02.g086200.t1.2 |
| Cre03.g149150.t1.2 | Cre03.g173000.t1.1 |
| Cre03.g179750.t1.2 | Cre05.g239400.t1.1 |
| Cre06.g278147.t1.1 | Cre06.g278209.t2.1 CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein |
| Cre07.g332800.t1.2 | Cre09.g390726.t1.1 |
| Cre10.g436650.t1.1 DEA(D/H)-box RNA helicase family protein | Cre11.g467541.t1.1 |
| Cre11.g467740.t1.1 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Cre12.g519700.t1.1 P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| Cre12.g540200.t1.2 DEA(D/H)-box RNA helicase family protein | Cre13.g602300.t1.2 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| Cre14.g613200.t1.1 plastid transcriptionally active 17 | Cre16.g690767.t1.1 |
| Cre17.g716400.t1.2 Zinc finger protein 622 | Cre17.g722250.t1.1 |
|
e.value: 0.00028 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: GCagnnGCTGcTGcnGaTGgt |
motif_0123_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 3400 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CTCcGCCGCCGcCgcCgCcgC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

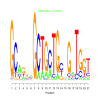

Comments