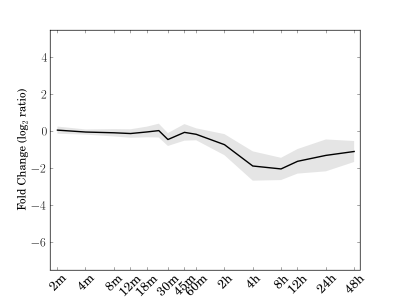
Module 211 Residual: 0.22
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0211 | v02 | 0.22 | -13.20 |
Displaying 1 - 21 of 21
| Cre02.g091450.t1.1 | Cre02.g100400.t1.2 |
| Cre02.g109600.t1.2 myo-inositol monophosphatase like 1 | Cre03.g156600.t1.2 proton gradient regulation 7 |
| Cre03.g184550.t1.2 Low PSII Accumulation 3 | Cre03.g207601.t1.1 |
| Cre04.g213100.t1.1 alpha/beta-Hydrolases superfamily protein | Cre05.g246800.t1.2 enzyme binding;tetrapyrrole binding |
| Cre07.g328150.t1.1 PDI-like 2-3 | Cre09.g416200.t1.2 high chlorophyll fluorescent 107 |
| Cre12.g500650.t1.2 Ribonuclease II/R family protein | Cre12.g530100.t1.2 Acid phosphatase/vanadium-dependent haloperoxidase-related protein |
| Cre13.g567950.t1.2 ADPGLC-PPase large subunit | Cre13.g572100.t1.1 |
| Cre13.g576760.t1.1 Chlorophyll A-B binding family protein | Cre13.g583400.t1.1 |
| Cre13.g606050.t1.2 Tyrosyl-tRNA synthetase, class Ib, bacterial/mitochondrial | Cre16.g656400.t1.2 sulfoquinovosyldiacylglycerol 1 |
| Cre16.g663900.t1.2 hydroxymethylbilane synthase | Cre16.g684350.t1.2 glyoxylate reductase 1 |
| Cre17.g736900.t1.2 |
|
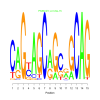
e.value: 0.0000000000000036 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: CAGcAGCaGcaGCAG |
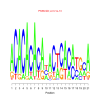
motif_0211_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:17e.value: 0.00000000000031 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: ACACACACAnACtcACatgcA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre12.g523000.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51zinc finger protein 1 |
0.055044 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

Comments