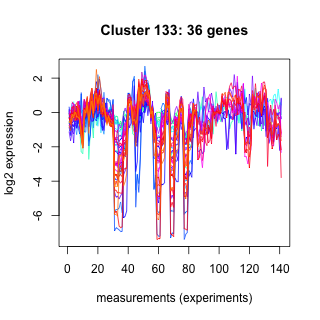
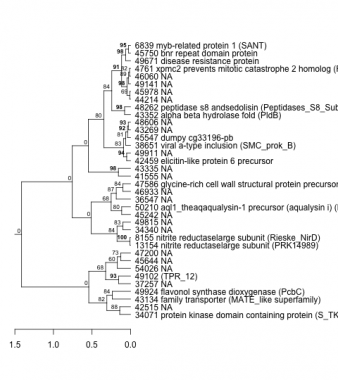
Phatr_hclust_0133 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006725 | aromatic compound metabolism | 0.0340932 | 1 | Phatr_hclust_0133 |
| GO:0006508 | proteolysis and peptidolysis | 0.053216 | 1 | Phatr_hclust_0133 |
| GO:0006118 | electron transport | 0.055391 | 1 | Phatr_hclust_0133 |
| GO:0006468 | protein amino acid phosphorylation | 0.262204 | 1 | Phatr_hclust_0133 |
| GO:0006810 | transport | 0.287164 | 1 | Phatr_hclust_0133 |
|
PHATRDRAFT_34071 : protein kinase domain containing protein (S_TKc) |
PHATRDRAFT_13154 : nitrite reductaselarge subunit (PRK14989) |
PHATRDRAFT_48262 : peptidase s8 andsedolisin (Peptidases_S8_Subtilisin_subset) |
|
|
PHATRDRAFT_8155 : nitrite reductaselarge subunit (Rieske_NirD) |
|||
|
PHATRDRAFT_43134 : family transporter (MATE_like superfamily) |
PHATRDRAFT_42459 : elicitin-like protein 6 precursor |
||
|
PHATRDRAFT_49924 : flavonol synthase dioxygenase (PcbC) |
|||
|
PHATRDRAFT_38651 : viral a-type inclusion (SMC_prok_B) |
|||
|
PHATRDRAFT_49102 : (TPR_12) |
PHATRDRAFT_50210 : aql1_theaqaqualysin-1 precursor (aqualysin i) (Peptidases_S8_PCSK9_ProteinaseK_like) |
PHATRDRAFT_45547 : dumpy cg33196-pb |
PHATRDRAFT_4761 : xpmc2 prevents mitotic catastrophe 2 homolog (REX4_like) |
|
PHATRDRAFT_49671 : disease resistance protein |
|||
|
PHATRDRAFT_45750 : bnr repeat domain protein |
|||
|
PHATRDRAFT_47586 : glycine-rich cell wall structural protein precursor (partial match) |
PHATRDRAFT_43352 : alpha beta hydrolase fold (PldB) |
PHATRDRAFT_6839 : myb-related protein 1 (SANT) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | -1.571 | 0.000505051 |
| Copper_SH | Copper_SH | 0.339 | 0.000515464 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.294 | 0.00178571 |
| Silver_SH | Silver_SH | -0.703 | 0.000555556 |
| Cadmium_1.2mg | Cadmium_1.2mg | -0.261 | 0.00104167 |
| BlueLight_24h | BlueLight_24h | -0.047 | 0.885484 |
| GreenLight_0.5h | GreenLight_0.5h | -2.894 | 0.000458716 |
| RedLight_6h | RedLight_6h | -0.989 | 0.000694444 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.980594 |
| GreenLight_24h | GreenLight_24h | 0.020 | 0.934233 |
| light_6hr | light_6hr | -0.856 | 0.000561798 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -1.217 | 0.00060241 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | 0.162 | 0.0290541 |
| Ammonia_SH | Ammonia_SH | -1.121 | 0.000344828 |
| Simazine_SH | Simazine_SH | -0.458 | 0.000423729 |
| Oil_SH | Oil_SH | 0.442 | 0.000666667 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.449 | 0.000609756 |
| Cadmium_SH | Cadmium_SH | -0.408 | 0.000666667 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.361 | 0.00777778 |
| highlight_0to6h | highlight_0to6h | -0.225 | 0.0156863 |
| BlueLight_0.5h | BlueLight_0.5h | -3.049 | 0.000462963 |
| Dispersant_SH | Dispersant_SH | 0.336 | 0.00298508 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | 0.848 | 0.000943396 |
| light_15.5hr | light_15.5hr | 1.046 | 0.000625 |
| light_10.5hr | light_10.5hr | 0.381 | 0.0571809 |
| Dispersed_oil_SH | Dispersed_oil_SH | 0.392 | 0.000675676 |
| Si_free | Si_free | -0.265 | 0.056 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | -2.766 | 0.000396825 |
| Mixture_SH | Mixture_SH | -1.155 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.151 | 0.368011 |
| light_16hr_dark_30min | light_16hr_dark_30min | 1.161 | 0.000735294 |
| BlueLight_6h | BlueLight_6h | -0.838 | 0.000581395 |
| highlight_12to48h | highlight_12to48h | 0.385 | 0.00384615 |
| RedLight_24h | RedLight_24h | 0.314 | 0.00952381 |
| Dark_treated | Dark_treated | -2.767 | 0.000446429 |
| GreenLight_6h | GreenLight_6h | -1.000 | 0.000609756 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.111 | 0.00185185 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.067 | 0.289045 |
| RedLight_0.5h | RedLight_0.5h | -2.930 | 0.000555556 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.010 | 0.959158 |
| Re-illuminated_24h | Re-illuminated_24h | 0.297 | 0.00119048 |