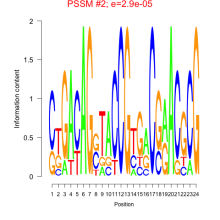
Phatr_bicluster_0134 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0134 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12252 n-ethylmaleimide sensitive fusion (RPT1)
PHATRDRAFT_12740 amidase family protein (Amidase superfamily)
PHATRDRAFT_14549 protein tyrosine (DSPc superfamily)
PHATRDRAFT_16052 glycoside hydrolase family 28 protein polygalacturonasefamily protein (Pectate_lyase_3 superfamily)
PHATRDRAFT_23706 proteolipid c subunit (ATP-synt_C superfamily)
PHATRDRAFT_30466 nad-dependent epimerase dehydratase (NADB_Rossmann superfamily)
PHATRDRAFT_36435 flavin reductase (NADB_Rossmann superfamily)
PHATRDRAFT_37711 (Lipase superfamily)
PHATRDRAFT_41170 quinol-to-oxygen oxidoreductase (Ferritin_like superfamily)
PHATRDRAFT_41947 fad dependent oxidoreductase (PRK11728 superfamily)
PHATRDRAFT_45084 (TPR)
PHATRDRAFT_49391 amino acid transporter (Aa_trans superfamily)
PHATRDRAFT_49879 ac034107_6ests gb
PHATRDRAFT_54153 ama1 protein (PLAC8)
PHATRDRAFT_7337 (Got1)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006629 | lipid metabolism | 0.0307293 | 1 | Phatr_bicluster_0134 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0324125 | 1 | Phatr_bicluster_0134 |
| GO:0006865 | amino acid transport | 0.0474487 | 1 | Phatr_bicluster_0134 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0622842 | 1 | Phatr_bicluster_0134 |
| GO:0006810 | transport | 0.287164 | 1 | Phatr_bicluster_0134 |
| GO:0006118 | electron transport | 0.33492 | 1 | Phatr_bicluster_0134 |
| GO:0008152 | metabolism | 0.410263 | 1 | Phatr_bicluster_0134 |

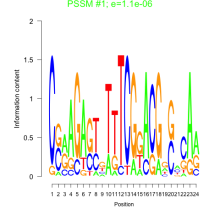
Comments