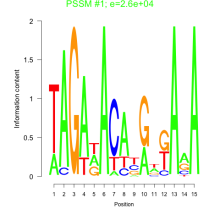
Phatr_bicluster_0158 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0158 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_16379 myosin heavy chain-like protein (MYSc)
PHATRDRAFT_38879 tetratricopeptide tpr_2 (TPR_12)
PHATRDRAFT_44214 (PHA03247)
PHATRDRAFT_47587 glycine-rich cell wall structural protein precursor (partial match)
PHATRDRAFT_47727 hypothetical glycine-rich protein
PHATRDRAFT_48061 (UBQ superfamily)
PHATRDRAFT_48544 gtpase activator protein (RabGAP-TBC superfamily)
PHATRDRAFT_49838 viral a-type inclusion (ANK)
PHATRDRAFT_49911 myb-like dna-bindingshaqkyf class family protein (SANT)
PHATRDRAFT_6311 importin alpha 1b (ARM)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006464 | protein modification | 0.0155594 | 1 | Phatr_bicluster_0158 |


Comments