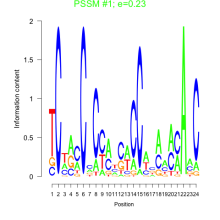
Thaps_bicluster_0133 Residual: 0.40
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0133 | 0.40 | Thalassiosira pseudonana |
Displaying 1 - 31 of 31
" class="views-fluidgrid-wrapper clear-block">
10052 hypothetical protein
11432 hypothetical protein
14106 LrgB superfamily
2012 hypothetical protein
22405 Delta6-FADS-like
22768 hypothetical protein
2344 hypothetical protein
24413 LCAT
25040 NA
261688 hypothetical protein
264121 GST_N_Sigma_like
268615 COG0579
270253 (NTT8) hypothetical protein
2991 (Tp_CCCH22) DnaQ_like_exo superfamily
30004 COG0385
31394 COG0436
3202 hypothetical protein
38800 plant_peroxidase_like superfamily
41005 odpB
4424 LabA_like
4875 PRK06247
5021 fabF
5175 Flavokinase
5996 N2227 superfamily
6028 NA
6253 hypothetical protein
711 COG0385
7155 hypothetical protein
7677 (DDE) diadinoxanthin de-epoxidase
ThpsCp123 (tufA) elongation factor Tu
ThpsCt015 (trnfM(cau))
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006814 | sodium ion transport | 0.000255764 | 0.698747 | Thaps_bicluster_0133 |
| GO:0009231 | riboflavin biosynthesis | 0.0135745 | 1 | Thaps_bicluster_0133 |
| GO:0006636 | fatty acid desaturation | 0.0269812 | 1 | Thaps_bicluster_0133 |
| GO:0006633 | fatty acid biosynthesis | 0.0424129 | 1 | Thaps_bicluster_0133 |
| GO:0006979 | response to oxidative stress | 0.053299 | 1 | Thaps_bicluster_0133 |
| GO:0006629 | lipid metabolism | 0.0640724 | 1 | Thaps_bicluster_0133 |
| GO:0006096 | glycolysis | 0.0810783 | 1 | Thaps_bicluster_0133 |
| GO:0009058 | biosynthesis | 0.118319 | 1 | Thaps_bicluster_0133 |
| GO:0006412 | protein biosynthesis | 0.319237 | 1 | Thaps_bicluster_0133 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0133 |

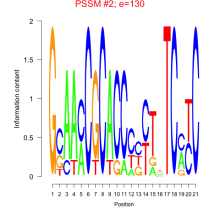
Comments