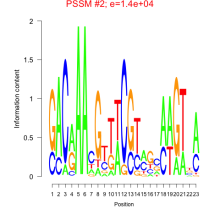
Thaps_bicluster_0177 Residual: 0.33
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0177 | 0.33 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
1082 hypothetical protein
11639 hypothetical protein
20933 hypothetical protein
21563 hypothetical protein
22763 DUF525 superfamily
23013 Trs65
24227 RNR_PFL superfamily
262412 Ptr
264143 PP2Cc
26702 ATPase-Plipid
268446 (CLPC1) clpA
31551 Aminotran_1_2
3874 hypothetical protein
5246 hypothetical protein
5659 hypothetical protein
5902 hypothetical protein
6200 hypothetical protein
7188 PKc_like superfamily
7546 COG1505
7937 CHD
8775 DAP2
9425 TPR_12
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007275 | development | 0.00495611 | 1 | Thaps_bicluster_0177 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0532936 | 1 | Thaps_bicluster_0177 |
| GO:0007165 | signal transduction | 0.0949469 | 1 | Thaps_bicluster_0177 |
| GO:0007242 | intracellular signaling cascade | 0.0994698 | 1 | Thaps_bicluster_0177 |
| GO:0006812 | cation transport | 0.101723 | 1 | Thaps_bicluster_0177 |
| GO:0019538 | protein metabolism | 0.108453 | 1 | Thaps_bicluster_0177 |
| GO:0009058 | biosynthesis | 0.128367 | 1 | Thaps_bicluster_0177 |
| GO:0006508 | proteolysis and peptidolysis | 0.157438 | 1 | Thaps_bicluster_0177 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.174355 | 1 | Thaps_bicluster_0177 |
| GO:0006468 | protein amino acid phosphorylation | 0.41786 | 1 | Thaps_bicluster_0177 |


Comments