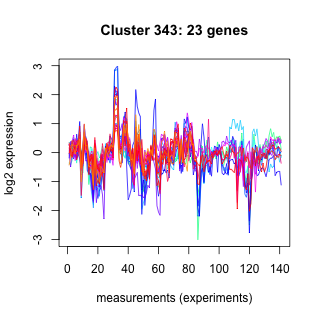
Phatr_hclust_0343 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0030163 | protein catabolism | 0.0242101 | 1 | Phatr_hclust_0343 |
| GO:0006259 | DNA metabolism | 0.026385 | 1 | Phatr_hclust_0343 |
| GO:0006306 | DNA methylation | 0.0436294 | 1 | Phatr_hclust_0343 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0457657 | 1 | Phatr_hclust_0343 |
| GO:0006508 | proteolysis and peptidolysis | 0.0848415 | 1 | Phatr_hclust_0343 |
| GO:0006412 | protein biosynthesis | 0.306181 | 1 | Phatr_hclust_0343 |
| GO:0006810 | transport | 0.352949 | 1 | Phatr_hclust_0343 |
| GO:0008152 | metabolism | 0.492943 | 1 | Phatr_hclust_0343 |
|
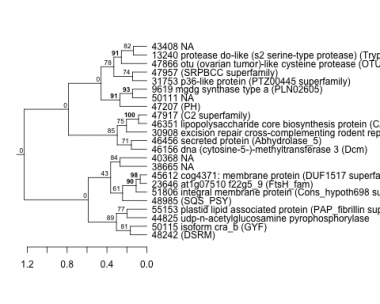
PHATRDRAFT_48242 : (DSRM) |
PHATRDRAFT_23646 : at1g07510 f22g5_9 (FtsH_fam) |
PHATRDRAFT_30908 : excision repair cross-complementing rodent repaircomplementation group 4 (rad1) |
PHATRDRAFT_31753 : p36-like protein (PTZ00445 superfamily) |
|
PHATRDRAFT_50115 : isoform cra_b (GYF) |
PHATRDRAFT_45612 : cog4371: membrane protein (DUF1517 superfamily) |
PHATRDRAFT_46351 : lipopolysaccharide core biosynthesis protein (CAP10 superfamily) |
PHATRDRAFT_47957 : (SRPBCC superfamily) |
|
PHATRDRAFT_44825 : udp-n-acetylglucosamine pyrophosphorylase |
PHATRDRAFT_47917 : (C2 superfamily) |
PHATRDRAFT_47866 : otu (ovarian tumor)-like cysteine protease (OTU) |
|
|
PHATRDRAFT_55153 : plastid lipid associated protein (PAP_fibrillin superfamily) |
PHATRDRAFT_47207 : (PH) |
PHATRDRAFT_13240 : protease do-like (s2 serine-type protease) (Trypsin_2) |
|
|
PHATRDRAFT_48985 : (SQS_PSY) |
PHATRDRAFT_46156 : dna (cytosine-5-)-methyltransferase 3 (Dcm) |
||
|
PHATRDRAFT_51806 : integral membrane protein (Cons_hypoth698 superfamily) |
PHATRDRAFT_46456 : secreted protein (Abhydrolase_5) |
PHATRDRAFT_9619 : mgdg synthase type a (PLN02605) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.605 | 0.000505051 |
| Copper_SH | Copper_SH | -0.184 | 0.0669811 |
| Green_vs_Red_24h | Green_vs_Red_24h | 0.055 | 0.653488 |
| Silver_SH | Silver_SH | 0.099 | 0.284682 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.096 | 0.194152 |
| BlueLight_24h | BlueLight_24h | -0.204 | 0.155714 |
| GreenLight_0.5h | GreenLight_0.5h | -0.033 | 0.944283 |
| RedLight_6h | RedLight_6h | 0.059 | 0.809637 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | -0.124 | 0.430843 |
| light_6hr | light_6hr | 0.281 | 0.214407 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.361 | 0.0673469 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.018 | 0.890233 |
| Ammonia_SH | Ammonia_SH | -0.672 | 0.013785 |
| Simazine_SH | Simazine_SH | 0.078 | 0.49631 |
| Oil_SH | Oil_SH | -0.128 | 0.206677 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.082 | 0.44963 |
| Cadmium_SH | Cadmium_SH | -0.150 | 0.189039 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.024 | 0.893356 |
| highlight_0to6h | highlight_0to6h | -0.013 | 0.967089 |
| BlueLight_0.5h | BlueLight_0.5h | 0.370 | 0.437234 |
| Dispersant_SH | Dispersant_SH | 0.144 | 0.167424 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.366 | 0.00886076 |
| light_15.5hr | light_15.5hr | -0.689 | 0.00121951 |
| light_10.5hr | light_10.5hr | 0.071 | 0.810747 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.215 | 0.0566514 |
| Si_free | Si_free | 0.084 | 0.480702 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 0.248 | 0.607377 |
| Mixture_SH | Mixture_SH | -0.870 | 0.00208333 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.306 | 0.107576 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.645 | 0.00454545 |
| BlueLight_6h | BlueLight_6h | 0.365 | 0.021978 |
| highlight_12to48h | highlight_12to48h | -0.585 | 0.000555556 |
| RedLight_24h | RedLight_24h | -0.179 | 0.248927 |
| Dark_treated | Dark_treated | 1.633 | 0.0018797 |
| GreenLight_6h | GreenLight_6h | 0.383 | 0.023262 |
| Cadmium_0.12mg | Cadmium_0.12mg | 0.002 | 0.982813 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.079 | 0.306354 |
| RedLight_0.5h | RedLight_0.5h | -0.607 | 0.0894472 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.324 | 0.0735955 |
| Re-illuminated_24h | Re-illuminated_24h | -0.106 | 0.494348 |