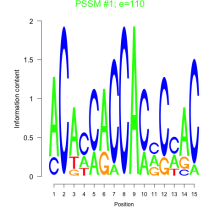
Thaps_bicluster_0155 Residual: 0.30
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0155 | 0.30 | Thalassiosira pseudonana |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
11643 (Tp_AP2-EREBP3) regulator [Rayko]
11799 hypothetical protein
11806 TRAM_LAG1_CLN8
11918 hypothetical protein
1224 MFS transporter
13175 COG4178
1968 hypothetical protein
2227 DUF3661
2376 PTZ00298
24220 hypothetical protein
24304 hypothetical protein
2497 hypothetical protein
261211 FGE-sulfatase superfamily
261327 NADB_Rossmann superfamily
262201 PP2Cc
30105 (FPS1) Trans_IPPS_HT
33794 ERG4_ERG24 superfamily
33926 p450 superfamily
34510 UAA
3750 hypothetical protein
7227 hypothetical protein
8587 hypothetical protein
9133 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008299 | isoprenoid biosynthesis | 0.0225308 | 1 | Thaps_bicluster_0155 |
| GO:0016310 | phosphorylation | 0.0265779 | 1 | Thaps_bicluster_0155 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0525183 | 1 | Thaps_bicluster_0155 |
| GO:0009058 | biosynthesis | 0.108157 | 1 | Thaps_bicluster_0155 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.129013 | 1 | Thaps_bicluster_0155 |
| GO:0008152 | metabolism | 0.203212 | 1 | Thaps_bicluster_0155 |
| GO:0006118 | electron transport | 0.432304 | 1 | Thaps_bicluster_0155 |
| GO:0006508 | proteolysis and peptidolysis | 0.459068 | 1 | Thaps_bicluster_0155 |


Comments