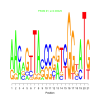
Module 28 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0028 | v02 | 0.52 | -17.50 |
| Rv0118c Oxalyl-CoA decarboxylase (EC 4.1.1.8) | Rv0119 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Rv0392c NADH dehydrogenase (EC 1.6.99.3) | Rv0393 13E12 repeat family protein |
| Rv1032c Sensor protein basS/pmrB (EC 2.7.3.-) | Rv1033c DNA-binding response regulator TrcR |
| Rv1034c transposase | Rv1375 |
| Rv1376 | Rv2026c Universal stress protein family |
| Rv2027c Histidine kinase response regulator | Rv2029c Tagatose-6-phosphate kinase (EC 2.7.1.144) / 1-phosphofructokinase (EC 2.7.1.56) |
| Rv2045c Probable carboxylesterase LipT (EC 3.1.1.-) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0007165 | GO:0023052 | GO:0044700 | GO:0007154 | GO:0050896 | GO:0004871 | GO:0060089 | GO:0000155 | GO:0004872 | GO:0038023 | signal transduction | signaling | single organism signaling | cell communication | response to stimulus | signal transducer activity | molecular transducer activity | phosphorelay sensor kinase activity | receptor activity | signaling receptor activity |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0003954 | NADH dehydrogenase activity |
|
GO:0003955 | NAD(P)H dehydrogenase (quinone) activity |
|
GO:0005829 | cytosol |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0004672 | protein kinase activity |
|
GO:0005509 | calcium ion binding |
|
GO:0030145 | manganese ion binding |
|
GO:0046777 | protein autophosphorylation |
|
GO:0008150 | biological_process |
|
GO:0003677 | DNA binding |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0045893 | positive regulation of transcription, DNA-dependent |
|
GO:0000287 | magnesium ion binding |
|
GO:0004673 | protein histidine kinase activity |
|
GO:0005524 | ATP binding |
|
GO:0019825 | oxygen binding |
|
GO:0019826 | oxygen sensor activity |
|
GO:0020037 | heme binding |
|
GO:0070025 | carbon monoxide binding |
|
GO:0070026 | nitric oxide binding |
|
GO:0070483 | detection of hypoxia |















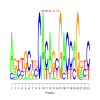
Discussion