Module 30 Residual: 0.60
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0030 | v02 | 0.60 | -17.77 |
| Rv0238 Transcriptional regulator, TetR family | Rv1303 ATP synthase protein I |
| Rv1304 ATP synthase A chain (EC 3.6.3.14) | Rv1305 ATP synthase C chain (EC 3.6.3.14) |
| Rv1306 ATP synthase B chain (EC 3.6.3.14) | Rv1307 ATP synthase B' chain (EC 3.6.3.14) / ATP synthase delta chain (EC 3.6.3.14) |
| Rv1308 ATP synthase alpha chain (EC 3.6.3.14) | Rv1309 ATP synthase gamma chain (EC 3.6.3.14) |
| Rv1310 ATP synthase beta chain (EC 3.6.3.14) | Rv1311 ATP synthase epsilon chain (EC 3.6.3.14) |
| Rv1312 Possible membrane protein | Rv1339 Metal-dependent hydrolases of the beta-lactamase superfamily III |
| Rv1863c Possible membrane protein | Rv2222c Glutamine synthetase type I (EC 6.3.1.2) |
| Rv3050c Transcriptional regulator, TetR family | Rv3677c FIG146518: Zn-dependent hydrolases, including glyoxylases |
| Rv3678c | Rv3679 Arsenical pump-driving ATPase (EC 3.6.3.16) |
| Rv3680 Arsenical pump-driving ATPase (EC 3.6.3.16) | Rv3704c Similar to Glutamate--cysteine ligase (EC 6.3.2.2), function unknown |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0044710 | GO:0044763 | GO:0009058 | GO:0034641 | GO:0006807 | GO:0044425 | single-organism metabolic process | single-organism cellular process | biosynthetic process | cellular nitrogen compound metabolic pro... | nitrogen compound metabolic process | membrane part |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0046961 | proton-transporting ATPase activity, rotational mechanism |
|
GO:0046933 | hydrogen ion transporting ATP synthase activity, rotational mechanism |
|
GO:0005887 | integral to plasma membrane |
|
GO:0005618 | cell wall |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0004356 | glutamate-ammonia ligase activity |
|
GO:0050001 | D-glutaminase activity |
|
GO:0051260 | protein homooligomerization |
|
GO:0008800 | beta-lactamase activity |
|
GO:0015446 | arsenite-transmembrane transporting ATPase activity |






















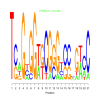
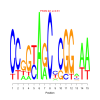
Discussion