Module 141 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0141 | v02 | 0.53 | -7.36 |
| Rv0109 PE-PGRS FAMILY PROTEIN | Rv0110 FIG056164: rhomboid family serine protease |
| Rv0262c Aminoglycoside 2'-N-acetyltransferase AAC (AAC(2')-IC) | Rv0263c Allophanate hydrolase 2 subunit 2 (EC 3.5.1.54) |
| Rv0264c Allophanate hydrolase 2 subunit 1 (EC 3.5.1.54) | Rv0858c putative aminotransferase |
| Rv1104 Carboxylesterase (EC 3.1.1.1) | Rv1434 |
| Rv2253 | Rv2176 Probable serine/threonine-protein kinase pknL (EC 2.7.11.1) |
| Rv3566c Arylamine N-acetyltransferase (EC 2.3.1.5) | Rv3772 Biosynthetic Aromatic amino acid aminotransferase beta (EC 2.6.1.57) |
|
GO:0005886 | plasma membrane |
|
GO:0034069 | aminoglycoside N-acetyltransferase activity |
|
GO:0047921 | aminoglycoside 2'-N-acetyltransferase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0005576 | extracellular region |
|
GO:0004672 | protein kinase activity |
|
GO:0004674 | protein serine/threonine kinase activity |
|
GO:0018107 | peptidyl-threonine phosphorylation |
|
GO:0046777 | protein autophosphorylation |
|
GO:0004060 | arylamine N-acetyltransferase activity |














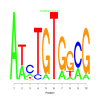

Discussion