Module 349 Residual: 0.51
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0349 | v02 | 0.51 | -4.40 |
| Rv0108c | Rv1248c Dihydrolipoamide succinyltransferase component (E2) of 2-oxoglutarate dehydrogenase complex (EC 2.3.1.61) / 2-oxoglutarate dehydrogenase E1 component (EC 1.2.4.2) |
| Rv1639c | Rv2302 |
| Rv2457c ATP-dependent Clp protease ATP-binding subunit ClpX | Rv3676 cAMP-binding proteins - catabolite gene activator and regulatory subunit of cAMP-dependent protein kinases |
| Rv0455c Possible membrane protein | Rv1182 POSSIBLE CONSERVED POLYKETIDE SYNTHASE ASSOCIATED PROTEIN PAPA2 |
| Rv2632c | Rv3119 Molybdenum cofactor biosynthesis protein MoaE; Molybdopterin converting factor subunit 2 |
| Rv3120 Possible methyltransferase (EC 2.1.1.-) | Rv3487c putative esterase |
| Rv3528c | Rv3588c Carbonic anhydrase (EC 4.2.1.1) |
| Rv3613c | Rv3614c ESX-1 secretion system protein (Rv3614c) |
| Rv3615c Highly immunodominant RD1 (Region of Difference 1)-dependent secreted antigen specific for Mycobacterium tuberculosis infection | Rv3616c ESX-1 secretion system protein (Rv3616c) |
| Rv3633 Protein involved in biosynthesis of mitomycin antibiotics/polyketide fumonisin | Rv3849 ESX-1 secreted protein regulator EspR |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0008653 | GO:0009103 | GO:0016830 | GO:0016833 | lipopolysaccharide metabolic process | lipopolysaccharide biosynthetic process | carbon-carbon lyase activity | oxo-acid-lyase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0005515 | protein binding |
|
GO:0005886 | plasma membrane |
|
GO:0006099 | tricarboxylic acid cycle |
|
GO:0008683 | 2-oxoglutarate decarboxylase activity |
|
GO:0009055 | electron carrier activity |
|
GO:0040007 | growth |
|
GO:0045254 | pyruvate dehydrogenase complex |
|
GO:0055114 | oxidation-reduction process |
|
GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity |
|
GO:0046983 | protein dimerization activity |
|
GO:0016887 | ATPase activity |
|
GO:0008270 | zinc ion binding |
|
GO:0019538 | protein metabolic process |
|
GO:0005618 | cell wall |
|
GO:0003677 | DNA binding |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0030552 | cAMP binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0045893 | positive regulation of transcription, DNA-dependent |
|
GO:0005576 | extracellular region |
|
GO:0004091 | carboxylesterase activity |
|
GO:0004629 | phospholipase C activity |
|
GO:0010447 | response to acidity |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0052572 | response to host immune response |
|
GO:0004089 | carbonate dehydratase activity |
|
GO:0051289 | protein homotetramerization |
|
GO:0009405 | pathogenesis |
|
GO:0042783 | active evasion of host immune response |
|
GO:0005829 | cytosol |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0050708 | regulation of protein secretion |






















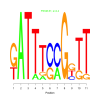

Discussion