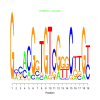
Module 364 Residual: 0.56
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0364 | v02 | 0.56 | -15.39 |
| Rv2054 Dienelactone hydrolase family protein | Rv2605c Acyl-CoA thioesterase II (EC 3.1.2.-) |
| Rv3042c Phosphoserine phosphatase (EC 3.1.3.3) | Rv3136 PPE family protein |
| Rv3210c | Rv3211 putative ATP-dependent RNA helicase |
| Rv1092c Pantothenate kinase (EC 2.7.1.33) | Rv2679 Enoyl-CoA hydratase (EC 4.2.1.17) |
| Rv3014c DNA ligase (EC 6.5.1.2) | Rv3038c Methyltransferase domain |
| Rv3039c Enoyl-CoA hydratase (EC 4.2.1.17) | Rv3040c |
| Rv3041c ABC transporter ATP-binding protein | Rv3198c ATP-dependent DNA helicase UvrD/PcrA, actinomycete paralog |
| Rv3212 Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage | Rv3216 Histone acetyltransferase HPA2 and related acetyltransferases |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0004647 | phosphoserine phosphatase activity |
|
GO:0040007 | growth |
|
GO:0004594 | pantothenate kinase activity |
|
GO:0004300 | enoyl-CoA hydratase activity |
|
GO:0003911 | DNA ligase (NAD+) activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0005618 | cell wall |
|
GO:0006266 | DNA ligation |
|
GO:0004003 | ATP-dependent DNA helicase activity |
|
GO:0005524 | ATP binding |
|
GO:0006281 | DNA repair |
|
GO:0003677 | DNA binding |
|
GO:0003678 | DNA helicase activity |
|
GO:0008094 | DNA-dependent ATPase activity |


















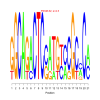
Discussion