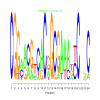
Module 388 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0388 | v02 | 0.53 | -17.30 |
| Rv0937c Ku domain protein | Rv0938 ATP-dependent DNA ligase (EC 6.5.1.1) clustered with Ku protein, LigD |
| Rv0939 Fumarylacetoacetase (EC 3.7.1.2) | Rv1011 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (EC 2.7.1.148) |
| Rv1166 LpqW | Rv1280c \Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein OppA (TC 3.A.1.5.1)\\\"" |
| Rv1562c Malto-oligosyltrehalose trehalohydrolase (EC 3.2.1.141) | Rv1563c Malto-oligosyltrehalose synthase (EC 5.4.99.15) |
| Rv1564c Glycogen debranching enzyme (EC 3.2.1.-) | Rv2364c GTP-binding protein Era |
| Rv0051 Multimodular transpeptidase-transglycosylase (EC 2.4.1.129) (EC 3.4.-.-) | Rv1281c Oligopeptide transport system permease protein OppD (TC 3.A.1.5.1) |
| Rv1282c Oligopeptide transport system permease protein OppC (TC 3.A.1.5.1) | Rv1283c Oligopeptide transport system permease protein OppB (TC 3.A.1.5.1) |
| Rv1522c Putative membrane protein MmpL12 | Rv1565c O-antigen acetylase |
| Rv1680 Phosphonate ABC transporter phosphate-binding periplasmic component (TC 3.A.1.9.1) | Rv1681 Molybdopterin biosynthesis protein MoeX |
|
GO:0003690 | double-stranded DNA binding |
|
GO:0005515 | protein binding |
|
GO:0006303 | double-strand break repair via nonhomologous end joining |
|
GO:0033202 | DNA helicase complex |
|
GO:0042803 | protein homodimerization activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0003887 | DNA-directed DNA polymerase activity |
|
GO:0003896 | DNA primase activity |
|
GO:0003899 | DNA-directed RNA polymerase activity |
|
GO:0003909 | DNA ligase activity |
|
GO:0003910 | DNA ligase (ATP) activity |
|
GO:0004652 | polynucleotide adenylyltransferase activity |
|
GO:0005524 | ATP binding |
|
GO:0005886 | plasma membrane |
|
GO:0006269 | DNA replication, synthesis of RNA primer |
|
GO:0008310 | single-stranded DNA specific 3'-5' exodeoxyribonuclease activity |
|
GO:0030145 | manganese ion binding |
|
GO:0004334 | fumarylacetoacetase activity |
|
GO:0005829 | cytosol |
|
GO:0050515 | 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase activity |
|
GO:0040007 | growth |
|
GO:0005576 | extracellular region |
|
GO:0009247 | glycolipid biosynthetic process |
|
GO:0005215 | transporter activity |
|
GO:0006810 | transport |
|
GO:0005887 | integral to plasma membrane |
|
GO:0033942 | 4-alpha-D-{(1->4)-alpha-D-glucano}trehalose trehalohydrolase activity |
|
GO:0047470 | (1,4)-alpha-D-glucan 1-alpha-D-glucosylmutase activity |
|
GO:0005975 | carbohydrate metabolic process |
|
GO:0043169 | cation binding |
|
GO:0003824 | catalytic activity |
|
GO:0003676 | nucleic acid binding |
|
GO:0005622 | intracellular |
|
GO:0005525 | GTP binding |
|
GO:0016020 | membrane |
|
GO:0052572 | response to host immune response |




















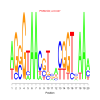
Discussion