Module 470 Residual: 0.49
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0470 | v02 | 0.49 | -8.09 |
| Rv1156 | Rv2373c Chaperone protein DnaJ |
| Rv3009c Aspartyl-tRNA(Asn) amidotransferase subunit B (EC 6.3.5.6) @ Glutamyl-tRNA(Gln) amidotransferase subunit B (EC 6.3.5.7) | Rv3430c Mobile element protein |
| Rv3581c 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (EC 4.6.1.12) | Rv3582c 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (EC 2.7.7.60) |
| Rv1178 N-succinyl-L,L-diaminopimelate aminotransferase alternative (EC 2.6.1.17) | Rv1354c Sensory box/GGDEF family protein |
| Rv1908c Catalase (EC 1.11.1.6) / Peroxidase (EC 1.11.1.7) | Rv3222c |
| Rv3223c RNA polymerase sigma-54 factor RpoN | Rv3346c |
|
GO:0031072 | heat shock protein binding |
|
GO:0006457 | protein folding |
|
GO:0051082 | unfolded protein binding |
|
GO:0040007 | growth |
|
GO:0010468 | regulation of gene expression |
|
GO:0005886 | plasma membrane |
|
GO:0005618 | cell wall |
|
GO:0008685 | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase activity |
|
GO:0050518 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0002135 | CTP binding |
|
GO:0008270 | zinc ion binding |
|
GO:0030145 | manganese ion binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0009016 | succinyldiaminopimelate transaminase activity |
|
GO:0004096 | catalase activity |
|
GO:0004601 | peroxidase activity |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0006979 | response to oxidative stress |
|
GO:0016677 | oxidoreductase activity, acting on a heme group of donors, nitrogenous group as acceptor |
|
GO:0020037 | heme binding |
|
GO:0042744 | hydrogen peroxide catabolic process |
|
GO:0070402 | NADPH binding |
|
GO:0070404 | NADH binding |
|
GO:0005515 | protein binding |
|
GO:0006950 | response to stress |
|
GO:0009408 | response to heat |
|
GO:0034605 | cellular response to heat |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0051409 | response to nitrosative stress |
|
GO:0052572 | response to host immune response |















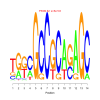
Discussion