Module 547 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0547 | v02 | 0.53 | -12.83 |
| Rv1027c DNA-binding response regulator KdpE | Rv1028c Osmosensitive K+ channel histidine kinase KdpD (EC 2.7.3.-) |
| Rv1029 Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1) | Rv1030 Potassium-transporting ATPase B chain (EC 3.6.3.12) (TC 3.A.3.7.1) |
| Rv1031 Potassium-transporting ATPase C chain (EC 3.6.3.12) (TC 3.A.3.7.1) | Rv2997 Phytoene dehydrogenase and related proteins |
| Rv0325 | Rv0329c |
| Rv0585c Possible membrane protein | Rv1141c Enoyl-CoA hydratase (EC 4.2.1.17) |
| Rv1357c Sensory box/GGDEF family protein | Rv1553 Succinate dehydrogenase iron-sulfur protein (EC 1.3.99.1) |
| Rv1767 Possible carboxymuconolactone decarboxylase family protein (EC 4.1.1.44) | Rv1913 Fumarylacetoacetase (EC 3.7.1.2) |
| Rv2317 Inositol transport system permease protein | Rv2541 |
| Rv2893 POSSIBLE OXIDOREDUCTASE (EC 1.-.-.-) | Rv3428c Mobile element protein |
| Rv3667 Acetyl-coenzyme A synthetase (EC 6.2.1.1) |
|
GO:0008150 | biological_process |
|
GO:0005515 | protein binding |
|
GO:0040007 | growth |
|
GO:0000155 | two-component sensor activity |
|
GO:0016020 | membrane |
|
GO:0006950 | response to stress |
|
GO:0005524 | ATP binding |
|
GO:0007165 | signal transduction |
|
GO:0005737 | cytoplasm |
|
GO:0005886 | plasma membrane |
|
GO:0008556 | potassium-transporting ATPase activity |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0004300 | enoyl-CoA hydratase activity |
|
GO:0000104 | succinate dehydrogenase activity |
|
GO:0005829 | cytosol |
|
GO:0005576 | extracellular region |
|
GO:0003987 | acetate-CoA ligase activity |
|
GO:0005618 | cell wall |





















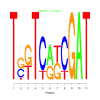
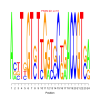
Discussion