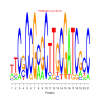
Module 31 Residual: 0.69
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0031 | v02 | 0.69 | -15.56 |
| Rv1296 Homoserine kinase (EC 2.7.1.39) | Rv1887 Possible membrane protein |
| Rv2674 Peptide methionine sulfoxide reductase MsrB (EC 1.8.4.12) | Rv2719c Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage |
| Rv2720 SOS-response repressor and protease LexA (EC 3.4.21.88) | Rv2737c RecA protein @ intein-containing |
| Rv2764c Thymidylate synthase (EC 2.1.1.45) | Rv2765 Dienelactone hydrolase family |
| Rv2846c EmrB/QacA family drug resistance transporter | Rv2917 DNA or RNA helicase of superfamily protein II |
| Rv2924c Formamidopyrimidine-DNA glycosylase (EC 3.2.2.23) | Rv2984 Polyphosphate kinase (EC 2.7.4.1) |
| Rv3711c DNA polymerase III epsilon subunit (EC 2.7.7.7) | Rv3712 proposed amino acid ligase found clustered with an amidotransferase |
| Rv3713 Putative amidotransferase similar to cobyric acid synthase |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0003684 | GO:0004197 | GO:0008234 | damaged DNA binding | cysteine-type endopeptidase activity | cysteine-type peptidase activity |
|
GO:0004413 | homoserine kinase activity |
|
GO:0040007 | growth |
|
GO:0005576 | extracellular region |
|
GO:0000318 | protein-methionine-R-oxide reductase activity |
|
GO:0006979 | response to oxidative stress |
|
GO:0051409 | response to nitrosative stress |
|
GO:0006974 | response to DNA damage stimulus |
|
GO:0016998 | cell wall macromolecule catabolic process |
|
GO:0046677 | response to antibiotic |
|
GO:0008992 | repressor LexA activity |
|
GO:0003677 | DNA binding |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0000150 | recombinase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0000725 | recombinational repair |
|
GO:0003697 | single-stranded DNA binding |
|
GO:0004520 | endodeoxyribonuclease activity |
|
GO:0005524 | ATP binding |
|
GO:0005829 | cytosol |
|
GO:0008094 | DNA-dependent ATPase activity |
|
GO:0009650 | UV protection |
|
GO:0016887 | ATPase activity |
|
GO:0030145 | manganese ion binding |
|
GO:0042148 | strand invasion |
|
GO:0004799 | thymidylate synthase activity |
|
GO:0005886 | plasma membrane |
|
GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity |
|
GO:0008976 | polyphosphate kinase activity |
|
GO:0001666 | response to hypoxia |
|
GO:0009405 | pathogenesis |
|
GO:0015968 | stringent response |
|
GO:0016310 | phosphorylation |
|
GO:0042803 | protein homodimerization activity |
|
GO:0046777 | protein autophosphorylation |
|
GO:0008763 | UDP-N-acetylmuramate-L-alanine ligase activity |

















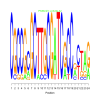
Discussion