MMP0304 MMP0304 conserved hypothetical archaeal protein (NCBI)
| Locus Tag | Symbol | Synonyms | Product | Protein Synonyms | Start | End | Strand | PubMed ID | |
|---|---|---|---|---|---|---|---|---|---|
| MMP0304 | MMP0304 | MMP0304 | MMP0304 | conserved hypothetical archaeal protein (NCBI) | N-glycosylase/DNA lyase (8-oxoguanine DNA glycosylase) (EC 3.2.2.-) (AGOG) (DNA-(apurinic or apyrimidinic site) lyase) (AP lyase) (EC 4.2.99.18) | 302428 | 301670 | - | 15466049 |
| Residual | Motif 1 | Expression Plot | |
|---|---|---|---|
| mmp-bicluster_0022 | 0.40 |
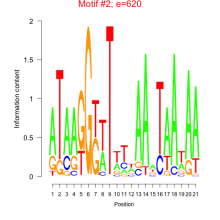 |
|
| mmp-bicluster_0033 | 0.21 |
 |
|
| mmp-bicluster_0043 | 0.30 |
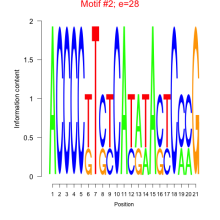 |
|
| mmp-bicluster_0152 | 0.25 |
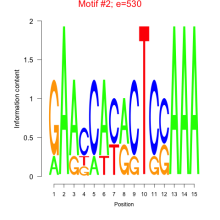 |
| Title | Insert | TAG Module |
|---|---|---|
| MMP0304 |
| Orthologues | Paralogues |
|---|---|
| Catalytic Activity | COG | EC Description | Protein families |
|---|---|---|---|
| The C-O-P bond 3' to the apurinic or apyrimidinic site in DNA is broken by a beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate. {ECO:0000255|HAMAP-Rule:MF_01168}. | (S) COG4047 | Uncharacterized protein conserved in archaea | (4.2.99.18) DNA-(apurinic or apyrimidinic site) lyase. | Archaeal N-glycosylase/DNA lyase (AGOG) family |
| L1.Cm.T1 | L1.Cm.T2 | L1.cN.T1 | L1.cN.T2 | L1.T0 | L2.Cm.T1 | L2.Cm.T2 | L2.cN.T1 | L2.cN.T2 | L2.T0 |
|---|---|---|---|---|---|---|---|---|---|
| 31 | 26 | 11 | 10 | 23 | 17 | 15 | 12 | 7 | 15 |
| Product (LegacyBRC) | Product (RefSeq) |
|---|---|
| GI Number | Accession | Blast | Conserved Domains |
|---|---|---|---|
| 45357867 | NP_987424.1 | Run |
| Link to STRINGS | STRINGS Network |
|---|---|
| MMP0304 |

Add comment