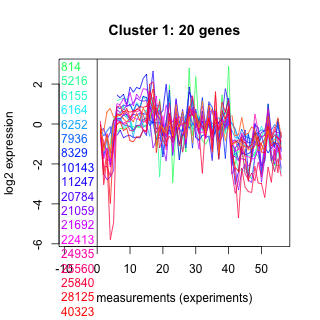
Thaps_hclust_0001 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006396 | RNA processing | 0.00377782 | 1 | Thaps_hclust_0001 |
| GO:0019856 | pyrimidine base biosynthesis | 0.00495457 | 1 | Thaps_hclust_0001 |
| GO:0006526 | arginine biosynthesis | 0.0147993 | 1 | Thaps_hclust_0001 |
| GO:0006807 | nitrogen compound metabolism | 0.0180619 | 1 | Thaps_hclust_0001 |
| GO:0006779 | porphyrin biosynthesis | 0.0180619 | 1 | Thaps_hclust_0001 |
| GO:0006306 | DNA methylation | 0.0470024 | 1 | Thaps_hclust_0001 |
| GO:0008152 | metabolism | 0.505245 | 1 | Thaps_hclust_0001 |
|
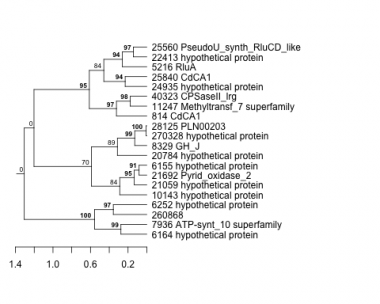
814 : CdCA1 |
7936 : ATP-synt_10 superfamily |
21059 : hypothetical protein |
25840 : CdCA1 |
|
5216 : RluA |
8329 : GH_J |
21692 : Pyrid_oxidase_2 |
28125 : PLN00203 |
|
6155 : hypothetical protein |
10143 : hypothetical protein |
22413 : hypothetical protein |
40323 : CPSaseII_lrg |
|
6164 : hypothetical protein |
11247 : Methyltransf_7 superfamily |
24935 : hypothetical protein |
260868 : |
|
6252 : hypothetical protein |
20784 : hypothetical protein |
25560 : PseudoU_synth_RluCD_like |
270328 : hypothetical protein |
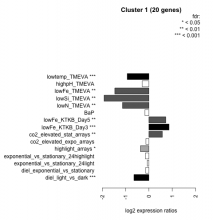
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.641 | 0.000485 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.863 | 0.000862 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.726 | 0.00132 |
| BaP | BaP | -0.171 | 0.467 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.150 | 0.163 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.578 | 0.00446 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.928 | 0.000735 |
| highpH_TMEVA | highpH_TMEVA | -0.289 | 0.0669 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.128 | 0.554 |
| lowFe_TMEVA | lowFe_TMEVA | -1.460 | 0.00104 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.069 | 0.867 |
| lowN_TMEVA | lowN_TMEVA | -1.130 | 0.00119 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.133 | 0.506 |
| lowSi_TMEVA | lowSi_TMEVA | -1.920 | 0.00135 |
| highlight_arrays | highlight_arrays | -0.362 | 0.0198 |