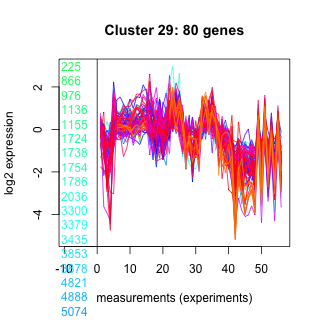
Thaps_hclust_0029 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006534 | cysteine metabolism | 0.00640893 | 1 | Thaps_hclust_0029 |
| GO:0006750 | glutathione biosynthetic process | 0.00640893 | 1 | Thaps_hclust_0029 |
| GO:0006457 | protein folding | 0.0089313 | 1 | Thaps_hclust_0029 |
| GO:0050757 | thymidylate synthase biosynthesis | 0.0191078 | 1 | Thaps_hclust_0029 |
| GO:0006783 | heme biosynthesis | 0.0253981 | 1 | Thaps_hclust_0029 |
| GO:0009765 | photosynthesis light harvesting | 0.0266581 | 1 | Thaps_hclust_0029 |
| GO:0009058 | biosynthesis | 0.0478395 | 1 | Thaps_hclust_0029 |
| GO:0006820 | anion transport | 0.0501715 | 1 | Thaps_hclust_0029 |
| GO:0006099 | tricarboxylic acid cycle | 0.0623285 | 1 | Thaps_hclust_0029 |
| GO:0008654 | phospholipid biosynthesis | 0.0861925 | 1 | Thaps_hclust_0029 |
|
225 : RRF |
6716 : CDP-OH_P_transf superfamily |
25361 : hypothetical protein |
262032 : hypothetical protein |
|
866 : S14_ClpP_2 |
6759 : hypothetical protein |
25787 : hypothetical protein |
262056 : (RS20) Ribosomal_S10 superfamily |
|
976 : hypothetical protein |
8085 : hypothetical protein |
26131 : PRK13474 |
263180 : SufS_like |
|
1136 : hypothetical protein |
10647 : hypothetical protein |
30981 : NADB_Rossmann superfamily |
264714 : Chloroa_b-bind superfamily |
|
1155 : hypothetical protein |
11739 : hypothetical protein |
31110 : (Tp_CSF2) CSP_CDS |
268621 : (PGM2) PLN02307 |
|
1724 : hypothetical protein |
11789 : hypothetical protein |
31128 : (lhcx5) Chloroa_b-bind |
268713 : hypothetical protein |
|
1738 : S14_ClpP_2 |
12030 : PRK05764 |
31216 : AAT_I superfamily |
270229 : hypothetical protein |
|
1754 : Gst |
13134 : AarF |
31402 : DUF3007 superfamily |
270314 : hypothetical protein |
|
1786 : hypothetical protein |
14192 : PLN02449 |
32845 : SDR_a5 |
270323 : hypothetical protein |
|
2036 : hypothetical protein |
17443 : IMPase |
33035 : YGGT |
ThpsCp020 : (psbN) photosystem II protein N |
|
3300 : hypothetical protein |
17704 : Peptidase_C12 superfamily |
33868 : EHD |
bd1545 : (bd1545) psbC |
|
3379 : hypothetical protein |
21042 : hypothetical protein |
34543 : (PPC2) PEPcase superfamily |
ThpsCp054 : (psbC) photosystem II 44 kDa protein |
|
3435 : Rotamase |
21250 : hypothetical protein |
34864 : FkpA |
bd1313 : (bd1313) psbH |
|
3853 : DUF3493 |
21300 : NADB_Rossmann superfamily |
36431 : cyclophilin_TLP40_like |
ThpsCp021 : (psbH) photosystem II protein H |
|
3878 : (PPI2) cyclophilin_ABH_like |
22527 : FNR_like |
38512 : Abhydrolase_6 |
ThpsCp019 : (psbT) photosystem II protein T |
|
4821 : hypothetical protein |
22689 : Chalcone superfamily |
38608 : SDR_a5 |
bd2073 : (bd2073) psbB |
|
4888 : hypothetical protein |
23278 : GrpE |
39841 : ArsA_ATPase |
ThpsCp018 : (psbB) photosystem II 47 kDa protein |
|
5074 : hypothetical protein |
23390 : hypothetical protein |
40193 : NA |
ThpsCp001 : (psaA) photosystem I P700 chlorophyll a apoprotein A1 |
|
5099 : DUF3119 superfamily |
24700 : hypothetical protein |
260962 : RHOD superfamily |
ThpsCp053 : (psbD) photosystem II protein D2 |
|
6564 : TspO_MBR superfamily |
24876 : hypothetical protein |
261284 : Aldo_ket_red |
ThpsCp067 : (petL) cytochrome b6/f complex subunit VI |
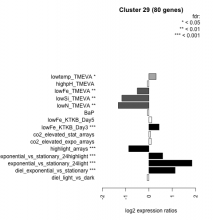
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.075 | 0.557 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.439 | 0.000862 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.119 | 0.24 |
| BaP | BaP | -0.062 | 0.612 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.591 | 0.000526 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.070 | 0.838 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.307 | 0.0152 |
| highpH_TMEVA | highpH_TMEVA | -0.065 | 0.555 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.101 | 0.487 |
| lowFe_TMEVA | lowFe_TMEVA | -0.500 | 0.00104 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 1.850 | 0.000581 |
| lowN_TMEVA | lowN_TMEVA | -1.310 | 0.00119 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 1.120 | 0.000602 |
| lowSi_TMEVA | lowSi_TMEVA | -1.160 | 0.00135 |
| highlight_arrays | highlight_arrays | -0.866 | 0.000442 |