Phatr_bicluster_0080 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0080 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11217 atp-dependent clp proteinase regulatory chain x (HslU)
PHATRDRAFT_11235 pentapeptide repeat
PHATRDRAFT_11733 mitochondrial creatine kinase (phosphagen_kinases superfamily)
PHATRDRAFT_2164 adp atp translocase (PTZ00169)
PHATRDRAFT_31434 mfs family transporter: sugar (galactose glucose) (Sugar_tr)
PHATRDRAFT_35678 imp protein (zf-UBP)
PHATRDRAFT_37440 rnase h family protein (RVT_3)
PHATRDRAFT_41886 chromosome 6 open reading frame 113 (Peptidase_C78 superfamily)
PHATRDRAFT_42422 hydroxysteroid (17-beta) dehydrogenase 10 (NADB_Rossmann superfamily)
PHATRDRAFT_42778 (DUF4239)
PHATRDRAFT_42971 (GLTP superfamily)
PHATRDRAFT_44278 (Cupin_8 superfamily)
PHATRDRAFT_44900 ubiquitin specific protease 22 (Peptidase_C19 superfamily)
PHATRDRAFT_45312 glycerophosphodiester phosphodiesterase (GDPD_GsGDE_like)
PHATRDRAFT_45381 transporter protein with conserved zn ribbon c11c7
PHATRDRAFT_45408 ntp pyrophosphohydrolase (Nudix_Hydrolase superfamily)
PHATRDRAFT_45563 glycine cleavage system regulatory protein (GcvR)
PHATRDRAFT_45576 (SEC14)
PHATRDRAFT_45611 cobyrinic acid-diamide synthase family protein
PHATRDRAFT_45973 s3 self-incompatibility locus-linked pollenprotein
PHATRDRAFT_46129 sec14 cytosolic factor family protein phosphoglyceride transfer family protein (SEC14)
PHATRDRAFT_9619 ubiquitin activating enzyme e1-like protein (E1_enzyme_family superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006071 | glycerol metabolism | 0.00788271 | 1 | Phatr_bicluster_0080 |
| GO:0008643 | carbohydrate transport | 0.0312056 | 1 | Phatr_bicluster_0080 |
| GO:0006810 | transport | 0.0513747 | 1 | Phatr_bicluster_0080 |
| GO:0015031 | protein transport | 0.0596835 | 1 | Phatr_bicluster_0080 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0855993 | 1 | Phatr_bicluster_0080 |
| GO:0008152 | metabolism | 0.109996 | 1 | Phatr_bicluster_0080 |


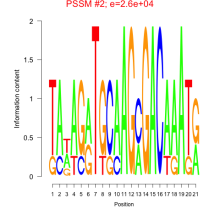
Comments