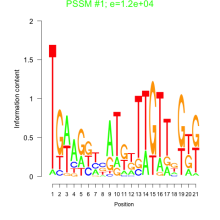
Thaps_bicluster_0052 Residual: 0.43
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0052 | 0.43 | Thalassiosira pseudonana |
Displaying 1 - 41 of 41
" class="views-fluidgrid-wrapper clear-block">
15267 COG1453
17039 MmsB
23184 MviM
23957 hypothetical protein
25812 Ribosomal_P0_L10e
262056 (RS20) Ribosomal_S10 superfamily
262257 YjgF_YER057c_UK114_like_2
269448 MATE_DinF_like
28704 Mtc superfamily
30981 NADB_Rossmann superfamily
6789 hypothetical protein
6932 hypothetical protein
7996 hypothetical protein
9605 hypothetical protein
ThpsCp005 (psbV) photosystem II cytochrome c550
ThpsCp006 (ycf66) hypothetical protein
ThpsCp007 (psbX) photosystem II protein X
ThpsCp010 (ycf24) putative ABC transporter
ThpsCp017 (atpA) ATP synthase CF1 alpha subunit
ThpsCp047 (ycf33) hypothetical protein
ThpsCp049 (petM) cytochrome b6-f complex subunit VII
ThpsCp057 (secG) preprotein translocase SecG subunit
ThpsCp072 (psbI) photosystem II protein I
ThpsCp083 (ycf46) Ycf46
ThpsCp087 (psaC) photosystem I subunit VII
ThpsCp088 (ccsA) cytochrome c biogenesis protein
ThpsCp092 (psbW) photosystem II protein W
ThpsCp129 (psaC) photosystem I subunit VII
ThpsCp133 (ycf46) Ycf46
ThpsCr002 (rnl)
ThpsCr005 (rnl')
ThpsCt001 (trnM(cau))
ThpsCt005 (trnF(gaa))
ThpsCt007 (trnT(ugu))
ThpsCt010 (trnD(guc))
ThpsCt012 (trnK(uuu))
ThpsCt018 (trnP(ugg))
ThpsCt023 (trnQ(uug))
ThpsCt025 (trnL(uaa))
ThpsCt027 (trnN(guu))
ThpsCt031 (trnP(ugg)')
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006414 | translational elongation | 0.000201196 | 0.550271 | Thaps_bicluster_0052 |
| GO:0006855 | multidrug transport | 0.0129601 | 1 | Thaps_bicluster_0052 |
| GO:0042254 | ribosome biogenesis and assembly | 0.0129601 | 1 | Thaps_bicluster_0052 |
| GO:0006812 | cation transport | 0.0772831 | 1 | Thaps_bicluster_0052 |
| GO:0008152 | metabolism | 0.171652 | 1 | Thaps_bicluster_0052 |
| GO:0006412 | protein biosynthesis | 0.269889 | 1 | Thaps_bicluster_0052 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0052 |

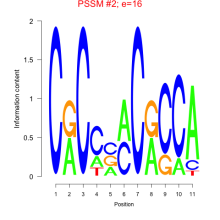
Comments