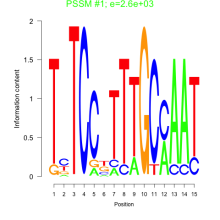
Thaps_bicluster_0059 Residual: 0.44
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0059 | 0.44 | Thalassiosira pseudonana |
Displaying 1 - 49 of 49
" class="views-fluidgrid-wrapper clear-block">
10541 ALDH-SF superfamily
10917 hypothetical protein
10996 hypothetical protein
11075 PRK12999
11076 PRK12999
11412 hypothetical protein
11636 COG1233
11676 hypothetical protein
11823 ANK
1341 hypothetical protein
14147 PRK08198
14242 S_TKc
1791 hypothetical protein
1855 hypothetical protein
2019 hypothetical protein
21045 hypothetical protein
21447 hypothetical protein
21539 EEVS
22372 HMGB-UBF_HMG-box
22409 hypothetical protein
23983 hypothetical protein
2420 RpsA
2521 Patatin_and_cPLA2 superfamily
261272 CBS_pair superfamily
261447 D-Amino acid dehydrogenase
263105 (FCL1) FAA1
263437 PLN02601 superfamily
264651 hypothetical protein
264759 Ggt
264846 Peptidase_C1A
268248 hypothetical protein
29728 NADB_Rossmann superfamily
3040 SRPBCC superfamily
31316 pyruv_ox_red
31979 ENDO3c
32944 PRK08139
38767 Peptidase_C1 superfamily
413 HIBADH
5330 hypothetical protein
5362 hypothetical protein
5500 (PDK1_2) PRK09279
6123 ANK
6361 PTZ00318
6531 hypothetical protein
6625 hypothetical protein
6873 PRK05463
7003 hypothetical protein
7325 hypothetical protein
9689 (Tp_bZIP7b/PAS) regulator [Rayko]
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006573 | valine metabolism | 0.000192527 | 0.526754 | Thaps_bicluster_0059 |
| GO:0006094 | gluconeogenesis | 0.00113463 | 1 | Thaps_bicluster_0059 |
| GO:0008152 | metabolism | 0.0262074 | 1 | Thaps_bicluster_0059 |
| GO:0006098 | pentose-phosphate shunt | 0.0342509 | 1 | Thaps_bicluster_0059 |
| GO:0006284 | base-excision repair | 0.0342509 | 1 | Thaps_bicluster_0059 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.0398483 | 1 | Thaps_bicluster_0059 |
| GO:0016117 | carotenoid biosynthesis | 0.0454144 | 1 | Thaps_bicluster_0059 |
| GO:0009225 | nucleotide-sugar metabolism | 0.0509494 | 1 | Thaps_bicluster_0059 |
| GO:0005975 | carbohydrate metabolism | 0.0692041 | 1 | Thaps_bicluster_0059 |
| GO:0016310 | phosphorylation | 0.0727814 | 1 | Thaps_bicluster_0059 |


Comments