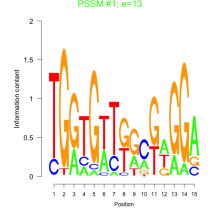
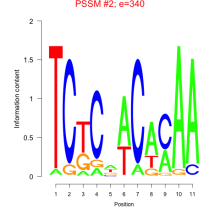
Thaps_bicluster_0066 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0066 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 34 of 34
" class="views-fluidgrid-wrapper clear-block">
14389 (HDC) PLN03032
17859 DER1 superfamily
20607 hypothetical protein
20983 DUF760
21291 hypothetical protein
21519 hypothetical protein
21600 Armet superfamily
2171 hypothetical protein
22547 hypothetical protein
2280 hypothetical protein
25605 (trpAB) Trp-synth_B
268061 hypothetical protein
30915 prmA
34340 hypothetical protein
34771 (MTF1) PLN02285
35780 rplC
3588 hypothetical protein
36456 eIF-5_eIF-2B superfamily
5514 NADB_Rossmann superfamily
6555 hypothetical protein
9049 hypothetical protein
9787 hypothetical protein
ThpsCp004 (rpl19) ribosomal protein L19
ThpsCp024 (rpl35) ribosomal protein L35
ThpsCp038 (ycf3) photosystem I assembly protein Ycf3
ThpsCp045 (rps14) ribosomal protein S14
ThpsCp079 (rpl21) ribosomal protein L21
ThpsCp080 (rpl27) ribosomal protein L27
ThpsCp095 (rpl3) ribosomal protein L3
ThpsCp126 (thiG) thiamin biosynthesis protein G
ThpsCp135 (secA) preprotein translocase subunit SecA
ThpsCp136 (rpl27) ribosomal protein L27
ThpsCp137 (rpl21) ribosomal protein L21
ThpsCt022 (trnH(gug))
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006568 | tryptophan metabolism | 0.0135745 | 1 | Thaps_bicluster_0066 |
| GO:0006310 | DNA recombination | 0.0424129 | 1 | Thaps_bicluster_0066 |
| GO:0006520 | amino acid metabolism | 0.0467809 | 1 | Thaps_bicluster_0066 |
| GO:0006413 | translational initiation | 0.053299 | 1 | Thaps_bicluster_0066 |
| GO:0006260 | DNA replication | 0.0768533 | 1 | Thaps_bicluster_0066 |
| GO:0006281 | DNA repair | 0.0998737 | 1 | Thaps_bicluster_0066 |
| GO:0009058 | biosynthesis | 0.118319 | 1 | Thaps_bicluster_0066 |
| GO:0008152 | metabolism | 0.235335 | 1 | Thaps_bicluster_0066 |
| GO:0006412 | protein biosynthesis | 0.319237 | 1 | Thaps_bicluster_0066 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0066 |

Comments