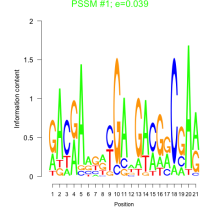
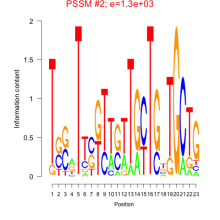
Thaps_bicluster_0082 Residual: 0.46
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0082 | 0.46 | Thalassiosira pseudonana |
Displaying 1 - 52 of 52
" class="views-fluidgrid-wrapper clear-block">
10460 hypothetical protein
10595 Rtt106
10661 hypothetical protein
1195 hypothetical protein
12160 hypothetical protein
12983 PKc_like superfamily
13343 Uup
1903 LRR_RI superfamily
19813 AdoMet_dc
20751 hypothetical protein
20909 hypothetical protein
21213 hypothetical protein
21316 hypothetical protein
21349 COG4857 superfamily
21392 hypothetical protein
22057 PI-PLCc_At5g67130_like
22495 hypothetical protein
22960 hypothetical protein
23335 hypothetical protein
23431 LRR_RI superfamily
23543 hypothetical protein
23796 hypothetical protein
23890 WcaG
24028 hypothetical protein
25095 hypothetical protein
25294 hypothetical protein
25476 OTU
25739 vWFA
260917 PDEase_I
262567 Aldo_ket_red
262924 PP2Cc
263461 arsM
263887 S_TKc
264297 MhpC
264671 S_TKc
269258 hypothetical protein
3255 hypothetical protein
32934 Rhomboid
33209 hypothetical protein
3614 hypothetical protein
36443 Peptidase_C14
36702 pyr_form_ly_1
38597 pyr_form_ly_1
3870 CIA30 superfamily
38760 pyr_form_ly_1
38845 ABCC_MRP_domain2
42545 PA superfamily
5456 hypothetical protein
5880 hypothetical protein
6743 hypothetical protein
7497 BCNT
9463 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006006 | glucose metabolism | 0.00000606 | 0.0167382 | Thaps_bicluster_0082 |
| GO:0006468 | protein amino acid phosphorylation | 0.0484723 | 1 | Thaps_bicluster_0082 |
| GO:0008152 | metabolism | 0.0696085 | 1 | Thaps_bicluster_0082 |
| GO:0006725 | aromatic compound metabolism | 0.0831178 | 1 | Thaps_bicluster_0082 |
| GO:0006508 | proteolysis and peptidolysis | 0.0999218 | 1 | Thaps_bicluster_0082 |
| GO:0007165 | signal transduction | 0.146211 | 1 | Thaps_bicluster_0082 |
| GO:0000074 | regulation of cell cycle | 0.211484 | 1 | Thaps_bicluster_0082 |
| GO:0016567 | protein ubiquitination | 0.269091 | 1 | Thaps_bicluster_0082 |
| GO:0006412 | protein biosynthesis | 0.485607 | 1 | Thaps_bicluster_0082 |
| GO:0006810 | transport | 0.52206 | 1 | Thaps_bicluster_0082 |

Comments