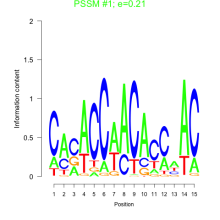
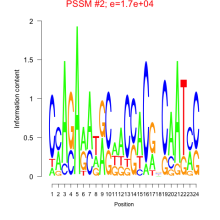
Thaps_bicluster_0147 Residual: 0.38
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0147 | 0.38 | Thalassiosira pseudonana |
Displaying 1 - 31 of 31
" class="views-fluidgrid-wrapper clear-block">
10556 ABC_ATPase superfamily
12259 COG1204
15246 NhaP
15336 DUS_like_FMN
1591 hypothetical protein
17271 FtsJ
19649 hypothetical protein
19743 Sm_D3
20800 XRN_N superfamily
21860 PRP40
22442 CHD
22757 (Tp_Hox6a) zf-CW
23095 UBCc
23096 hypothetical protein
23105 Peptidase_S74
23361 hypothetical protein
24288 RtcB
24942 COG3899
24980 hypothetical protein
25202 hypothetical protein
25222 hypothetical protein
25223 SNF2_N
25239 hypothetical protein
261258 (ZFP7) PHD
262126 Fe_hyd_lg_C superfamily
264322 MIF4G
269403 hypothetical protein
37824 Peptidase_C1A
3840 hypothetical protein
5776 PROCN superfamily
9591 efThoc1 superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0579953 | 1 | Thaps_bicluster_0147 |
| GO:0008033 | tRNA processing | 0.0579953 | 1 | Thaps_bicluster_0147 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0704475 | 1 | Thaps_bicluster_0147 |
| GO:0006397 | mRNA processing | 0.0704475 | 1 | Thaps_bicluster_0147 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0735363 | 1 | Thaps_bicluster_0147 |
| GO:0006306 | DNA methylation | 0.0918683 | 1 | Thaps_bicluster_0147 |
| GO:0006512 | ubiquitin cycle | 0.112826 | 1 | Thaps_bicluster_0147 |
| GO:0007242 | intracellular signaling cascade | 0.130427 | 1 | Thaps_bicluster_0147 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.14198 | 1 | Thaps_bicluster_0147 |
| GO:0006464 | protein modification | 0.178538 | 1 | Thaps_bicluster_0147 |

Comments