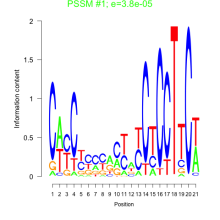
Thaps_bicluster_0233 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0233 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
10612 SseA
13572 (MTS1) PRK11893
20597 hypothetical protein
20739 hypothetical protein
20877 Ubiq_cyt_C_chap
21305 hypothetical protein
21804 hypothetical protein
21837 HSP33
21929 COG0116
22635 SIK1
23393 hypothetical protein
24550 W2_eIF4G1_like
24679 PCIF1_WW superfamily
25027 RRM_SF superfamily
261893 Ofd1_CTDD
262598 PLN00152
263880 PRK12267
269710 NADB_Rossmann superfamily
269901 spermidine synthase
29232 PLN02502
29782 (eIF6) IF6
3314 hypothetical protein
3416 TPR
354 PTZ00424
36045 UBCc
8157 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006351 | transcription, DNA-dependent | 0.00268762 | 1 | Thaps_bicluster_0233 |
| GO:0006431 | methionyl-tRNA aminoacylation | 0.00536856 | 1 | Thaps_bicluster_0233 |
| GO:0006430 | lysyl-tRNA aminoacylation | 0.00536856 | 1 | Thaps_bicluster_0233 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0097921 | 1 | Thaps_bicluster_0233 |
| GO:0006422 | aspartyl-tRNA aminoacylation | 0.0133715 | 1 | Thaps_bicluster_0233 |
| GO:0006413 | translational initiation | 0.0626925 | 1 | Thaps_bicluster_0233 |
| GO:0006306 | DNA methylation | 0.075288 | 1 | Thaps_bicluster_0233 |
| GO:0006512 | ubiquitin cycle | 0.0926592 | 1 | Thaps_bicluster_0233 |
| GO:0019538 | protein metabolism | 0.116953 | 1 | Thaps_bicluster_0233 |
| GO:0006464 | protein modification | 0.147638 | 1 | Thaps_bicluster_0233 |


Comments