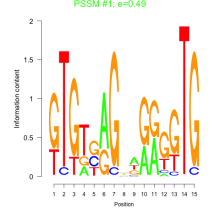
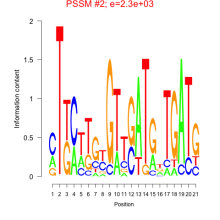
Thaps_bicluster_0268 Residual: 0.24
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0268 | 0.24 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
15992 COG0820
17292 Spb1_C superfamily
17654 REX4_like
20742 hypothetical protein
21966 SLX9
22334 hypothetical protein
22498 DEXDc superfamily
23202 RsuA
23277 EEP superfamily
261746 PRK00142
268486 Glyco_transf_22 superfamily
34729 NGP_1
39075 PRK09246
5294 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006396 | RNA processing | 0.00205534 | 1 | Thaps_bicluster_0268 |
| GO:0009113 | purine base biosynthesis | 0.00247959 | 1 | Thaps_bicluster_0268 |
| GO:0009116 | nucleoside metabolism | 0.00742341 | 1 | Thaps_bicluster_0268 |
| GO:0006118 | electron transport | 0.287914 | 1 | Thaps_bicluster_0268 |
| GO:0008152 | metabolism | 0.410014 | 1 | Thaps_bicluster_0268 |

Comments