Module 36 Residual: 0.21
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0036 | v02 | 0.21 | -13.07 |
Displaying 1 - 21 of 21
| Cre02.g074900.t1.1 Calcium-dependent lipid-binding (CaLB domain) family protein | Cre02.g105150.t1.1 |
| Cre02.g147900.t3.1 Pyruvate kinase family protein | Cre02.g147900.t4.1 Pyruvate kinase family protein |
| Cre03.g143887.t1.1 Arginyl-tRNA synthetase, class Ic | Cre03.g204250.t1.2 S-adenosyl-L-homocysteine hydrolase |
| Cre03.g206600.t1.2 dehydratase family | Cre04.g213761.t2.1 Thioesterase superfamily protein |
| Cre05.g233305.t1.1 NAD(P)-binding Rossmann-fold superfamily protein | Cre05.g245900.t1.2 branched-chain amino acid aminotransferase 5 / branched-chain amino acid transaminase 5 (BCAT5) |
| Cre06.g261150.t1.2 Radical SAM superfamily protein | Cre06.g273050.t1.2 Arabinanase/levansucrase/invertase |
| Cre08.g370650.t1.2 6-phosphogluconolactonase 1 | Cre08.g380000.t1.2 |
| Cre09.g395650.t1.1 | Cre09.g396250.t1.2 vitamin E pathway gene 5 |
| Cre10.g451900.t1.1 Pyridoxal-5\'-phosphate-dependent enzyme family protein | Cre12.g501550.t1.2 |
| Cre13.g566250.t1.2 | Cre14.g625750.t2.1 Tic22-like family protein |
| Cre16.g651750.t1.2 tRNA synthetase class I (I, L, M and V) family protein |
|
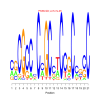
e.value: 0.000000085 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: CTgCTgctgCTGcTG |
motif_0036_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000057 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: cgCGCCcCcGCctCCGCcgcC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006096 | GO:0000287 | glycolysis | magnesium ion binding |



Comments