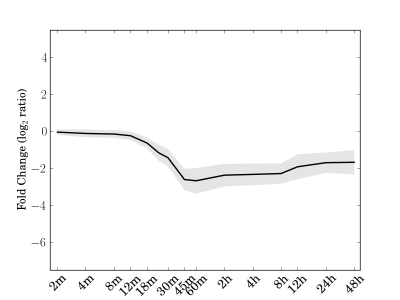
Module 44 Residual: 0.14
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0044 | v02 | 0.14 | -17.78 |
Displaying 1 - 21 of 21
| Cre01.g013100.t1.1 pathogenesis-related gene 1 | Cre01.g016300.t1.1 calmodulin-like 38 |
| Cre02.g080250.t1.2 YGGT family protein | Cre03.g148000.t1.2 |
| Cre03.g201750.t1.2 | Cre07.g352300.t1.2 |
| Cre08.g364450.t1.2 Acyl-CoA N-acyltransferases (NAT) superfamily protein | Cre08.g368700.t1.1 Rubisco methyltransferase family protein |
| Cre08.g375900.t1.1 Eukaryotic translation initiation factor eIF2A family protein | Cre08.g379650.t1.2 translocon at the inner envelope membrane of chloroplasts 20 |
| Cre08.g380201.t1.1 dehydroquinate dehydratase, putative / shikimate dehydrogenase, putative | Cre09.g386200.t1.1 |
| Cre09.g400950.t1.2 phosphate transporter 4;2 | Cre10.g430501.t1.1 Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein / tetratricopeptide repeat (TPR)-containing protein |
| Cre10.g441550.t1.1 CBS domain-containing protein with a domain of unknown function (DUF21) | Cre11.g467708.t2.1 |
| Cre12.g483550.t1.2 beta vacuolar processing enzyme | Cre12.g527550.t1.2 |
| Cre13.g576650.t1.2 histidine biosynthesis bifunctional protein (HISIE) | Cre14.g624850.t1.1 |
| Cre16.g696000.t1.2 N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
|
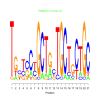
e.value: 4.5e-16 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: TgctcCtGCTgcTGCTgCTGC |
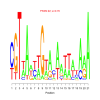
motif_0044_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.76 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: cGTtAnttGtacaattatAaA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0004635 | phosphoribosyl-AMP cyclohydrolase activi... |

Comments