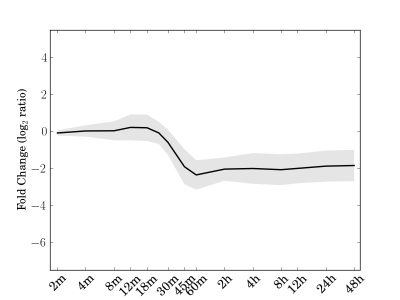
Module 139 Residual: 0.18
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0139 | v02 | 0.18 | -20.16 |
Displaying 1 - 20 of 20
| Cre01.g000150.t1.2 zinc transporter 7 precursor | Cre01.g002500.t1.2 Chlamyopsin, light-gated proton channel rhodopsin |
| Cre01.g049000.t1.2 Transcriptional coactivator/pterin dehydratase | Cre03.g198950.t1.2 Mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein |
| Cre06.g258733.t1.1 methylthioalkylmalate synthase-like 4 | Cre06.g308850.t1.2 Translation initiation factor eIF3 subunit |
| Cre07.g344350.t1.2 plastidic type i signal peptidase 1 | Cre07.g346050.t1.2 dicarboxylate diiron protein, putative (Crd1) |
| Cre08.g362100.t1.1 Similar to Flagellar Associated Protein FAP154 | Cre09.g389430.t1.1 phenylalanyl-tRNA synthetase, putative / phenylalanine--tRNA ligase, putative |
| Cre09.g391400.t1.1 | Cre09.g402100.t1.2 |
| Cre09.g409350.t1.2 RNAse l inhibitor protein 2 | Cre10.g434650.t1.1 Copper transport accessory protein |
| Cre12.g500950.t1.2 CLP protease P4 | Cre12.g516200.t1.2 Ribosomal protein S5/Elongation factor G/III/V family protein |
| Cre12.g532100.t1.2 | Cre16.g672385.t1.1 histidinol phosphate aminotransferase 1 |
| Cre16.g677000.t1.2 Heat shock protein 70 (Hsp 70) family protein | Cre17.g716950.t1.1 |
|
e.value: 9.2e-24 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: ACACAcaCaCACaCACacACACAC |
motif_0139_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.00000038 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: ACaCaCACACAcAtACACACA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |



Comments