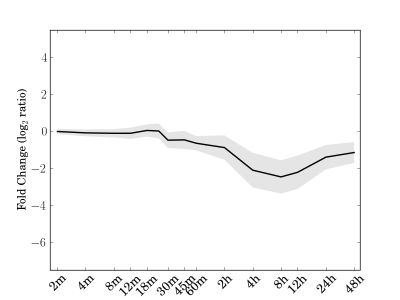
Module 146 Residual: 0.22
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0146 | v02 | 0.22 | -10.46 |
Displaying 1 - 21 of 21
| Cre01.g012150.t1.2 peptidemethionine sulfoxide reductase 3 | Cre01.g016500.t1.1 Dihydrolipoamide dehydrogenase |
| Cre01.g042800.t1.2 NAD(P)-binding Rossmann-fold superfamily protein | Cre02.g073700.t2.1 Uroporphyrinogen decarboxylase |
| Cre02.g091050.t1.2 Aldolase superfamily protein | Cre02.g109600.t1.2 myo-inositol monophosphatase like 1 |
| Cre02.g114450.t1.2 | Cre03.g201050.t1.2 thylakoid lumenal P17.1 protein |
| Cre06.g273700.t1.2 photosystem II stability/assembly factor, chloroplast (HCF136) | Cre06.g294750.t1.2 UbiA prenyltransferase family protein |
| Cre06.g300700.t1.1 NAD(P)-binding Rossmann-fold superfamily protein | Cre07.g328150.t1.1 PDI-like 2-3 |
| Cre09.g392579.t1.1 | Cre09.g405550.t1.2 Mitochondrial inner membrane translocase |
| Cre10.g429200.t1.1 Rubisco methyltransferase family protein | Cre12.g513750.t1.1 Glutaredoxin family protein |
| Cre12.g537850.t1.2 Protein of unknown function (DUF2930) | Cre12.g549350.t1.2 |
| Cre12.g554800.t1.2 phosphoribulokinase | Cre16.g684350.t1.2 glyoxylate reductase 1 |
| Cre17.g696350.t1.2 |
|
e.value: 0.00000000081 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: CAgCAGCAcCaGCAg |
motif_0146_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 7400 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: CAGCAAgcTGCAAGC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0033014 | tetrapyrrole biosynthetic process |

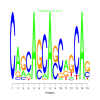

Comments