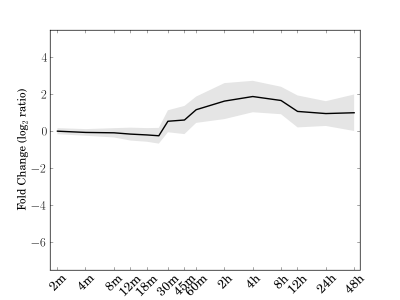
Module 198 Residual: 0.31
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0198 | v02 | 0.31 | -12.09 |
Displaying 1 - 30 of 30
| Cre01.g015250.t1.1 DNA binding;nucleotide binding;nucleic acid binding;DNA-directed DNA polymerases;DNA-directed DNA polymerases | Cre01.g047265.t1.1 |
| Cre02.g104100.t1.2 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | Cre03.g163150.t1.2 Putative glycosyl hydrolase of unknown function (DUF1680) |
| Cre03.g172900.t1.1 | Cre03.g183800.t1.1 |
| Cre03.g187850.t1.2 | Cre05.g247550.t1.2 glutathione S-transferase THETA 1 |
| Cre06.g260800.t1.2 protein binding | Cre06.g270550.t1.2 |
| Cre06.g277600.t1.2 | Cre06.g284150.t1.1 ammonium transporter 1;3 |
| Cre07.g320850.t1.2 beta glucosidase 42 | Cre07.g339300.t1.2 Zim17-type zinc finger protein |
| Cre08.g376350.t1.2 ASF1 like histone chaperone | Cre09.g386161.t1.1 |
| Cre09.g389950.t1.2 O-acetyltransferase family protein | Cre09.g410950.t1.2 nitrate reductase 2 |
| Cre09.g416050.t1.2 arginosuccinate synthase family | Cre10.g421300.t1.2 |
| Cre11.g467677.t1.1 RHOMBOID-like protein 13 | Cre11.g476650.t1.1 limit dextrinase |
| Cre12.g496550.t1.2 | Cre12.g528650.t1.2 |
| Cre13.g563100.t1.2 | Cre13.g584901.t1.1 |
| Cre13.g607500.t1.2 KU70 homolog | Cre14.g618150.t1.2 microtubule-associated proteins 70-2 |
| Cre16.g654050.t1.1 | Cre17.g739000.t2.1 |
|
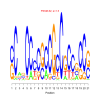
e.value: 0.0008 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: gcgGCAgcaGccgcgGCaGtgGCg |
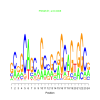
motif_0198_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 1.5 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: cCagCCcCgCCGCCgCCgCCg |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

Comments