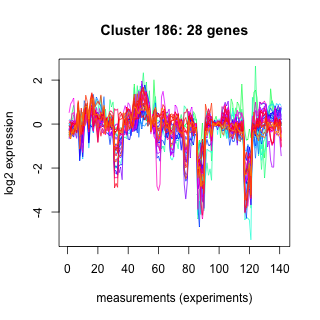
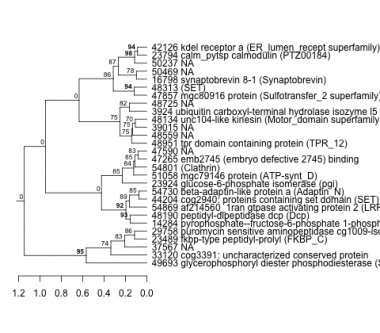
Phatr_hclust_0186 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006096 | glycolysis | 0.00453819 | 1 | Phatr_hclust_0186 |
| GO:0006094 | gluconeogenesis | 0.0147381 | 1 | Phatr_hclust_0186 |
| GO:0006508 | proteolysis and peptidolysis | 0.0253471 | 1 | Phatr_hclust_0186 |
| GO:0016192 | vesicle-mediated transport | 0.0379055 | 1 | Phatr_hclust_0186 |
| GO:0007018 | microtubule-based movement | 0.0854872 | 1 | Phatr_hclust_0186 |
| GO:0015031 | protein transport | 0.0882177 | 1 | Phatr_hclust_0186 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.104445 | 1 | Phatr_hclust_0186 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.125669 | 1 | Phatr_hclust_0186 |
| GO:0006457 | protein folding | 0.296964 | 1 | Phatr_hclust_0186 |
|
PHATRDRAFT_49693 : glycerophosphoryl diester phosphodiesterase (SdiA-regulated superfamily) |
PHATRDRAFT_54869 : af214560_1ran gtpase activating protein 2 (LRR_RI superfamily) |
PHATRDRAFT_47857 : mgc80916 protein (Sulfotransfer_2 superfamily) |
|
|
PHATRDRAFT_33120 : cog3391: uncharacterized conserved protein |
PHATRDRAFT_44204 : cog2940: proteins containing set domain (SET) |
PHATRDRAFT_48951 : tpr domain containing protein (TPR_12) |
PHATRDRAFT_48313 : (SET) |
|
PHATRDRAFT_54730 : beta-adaptin-like protein a (Adaptin_N) |
PHATRDRAFT_16798 : synaptobrevin 8-1 (Synaptobrevin) |
||
|
PHATRDRAFT_23489 : fkbp-type peptidyl-prolyl (FKBP_C) |
PHATRDRAFT_23924 : glucose-6-phosphate isomerase (pgi) |
||
|
PHATRDRAFT_29758 : puromycin sensitive aminopeptidase cg1009-isoform a (M1_APN_2) |
PHATRDRAFT_51058 : mgc79146 protein (ATP-synt_D) |
PHATRDRAFT_48134 : unc104-like kinesin (Motor_domain superfamily) |
|
|
PHATRDRAFT_14284 : pyrophosphate--fructose-6-phosphate 1-phosphotransferase (PFK superfamily) |
PHATRDRAFT_54801 : (Clathrin) |
PHATRDRAFT_3924 : ubiquitin carboxyl-terminal hydrolase isozyme l5 (Peptidase_C12_UCH37_BAP1) |
PHATRDRAFT_23794 : calm_pytsp calmodulin (PTZ00184) |
|
PHATRDRAFT_48190 : peptidyl-dipeptidase dcp (Dcp) |
PHATRDRAFT_47265 : emb2745 (embryo defective 2745) binding |
PHATRDRAFT_42126 : kdel receptor a (ER_lumen_recept superfamily) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.017 | 0.952376 |
| Copper_SH | Copper_SH | 0.174 | 0.0668561 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.118 | 0.279455 |
| Silver_SH | Silver_SH | 0.057 | 0.516953 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.020 | 0.826434 |
| BlueLight_24h | BlueLight_24h | -0.157 | 0.299185 |
| GreenLight_0.5h | GreenLight_0.5h | -0.572 | 0.0676744 |
| RedLight_6h | RedLight_6h | 0.097 | 0.657 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | -0.189 | 0.141333 |
| light_6hr | light_6hr | 0.413 | 0.0320856 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.220 | 0.234353 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.047 | 0.677333 |
| Ammonia_SH | Ammonia_SH | -2.204 | 0.000344828 |
| Simazine_SH | Simazine_SH | 0.148 | 0.162178 |
| Oil_SH | Oil_SH | -0.075 | 0.404737 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.004 | 0.973279 |
| Cadmium_SH | Cadmium_SH | -0.255 | 0.0210106 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.085 | 0.53949 |
| highlight_0to6h | highlight_0to6h | 0.143 | 0.31343 |
| BlueLight_0.5h | BlueLight_0.5h | -1.071 | 0.00604027 |
| Dispersant_SH | Dispersant_SH | -0.092 | 0.349519 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | 0.501 | 0.000943396 |
| light_15.5hr | light_15.5hr | 0.915 | 0.000625 |
| light_10.5hr | light_10.5hr | 0.714 | 0.00183486 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.145 | 0.14021 |
| Si_free | Si_free | -0.134 | 0.322897 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | -0.717 | 0.0463768 |
| Mixture_SH | Mixture_SH | -2.503 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.158 | 0.397772 |
| light_16hr_dark_30min | light_16hr_dark_30min | 0.857 | 0.000735294 |
| BlueLight_6h | BlueLight_6h | -0.061 | 0.72814 |
| highlight_12to48h | highlight_12to48h | 0.367 | 0.0240854 |
| RedLight_24h | RedLight_24h | -0.071 | 0.653571 |
| Dark_treated | Dark_treated | -1.126 | 0.0204023 |
| GreenLight_6h | GreenLight_6h | -0.015 | 0.936345 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.029 | 0.470349 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | 0.032 | 0.713942 |
| RedLight_0.5h | RedLight_0.5h | -0.527 | 0.107246 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.111 | 0.598381 |
| Re-illuminated_24h | Re-illuminated_24h | 0.176 | 0.147755 |