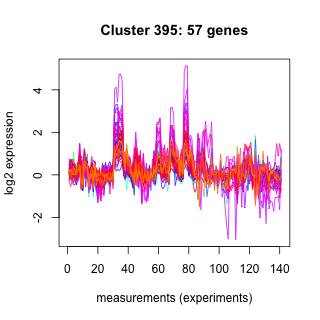
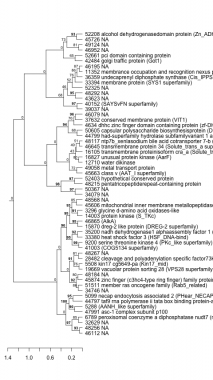
Phatr_hclust_0395 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006284 | base-excision repair | 0.00813004 | 1 | Phatr_hclust_0395 |
| GO:0006352 | transcription initiation | 0.032161 | 1 | Phatr_hclust_0395 |
| GO:0008152 | metabolism | 0.0403898 | 1 | Phatr_hclust_0395 |
| GO:0006468 | protein amino acid phosphorylation | 0.0767082 | 1 | Phatr_hclust_0395 |
| GO:0006508 | proteolysis and peptidolysis | 0.120633 | 1 | Phatr_hclust_0395 |
| GO:0016567 | protein ubiquitination | 0.17931 | 1 | Phatr_hclust_0395 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.45797 | 1 | Phatr_hclust_0395 |
|
PHATRDRAFT_28482 : cleavage and polyadenylation specific factor73kda (YSH1) |
PHATRDRAFT_48215 : pentatricopeptiderepeat-containing protein |
||
|
PHATRDRAFT_45663 : class v (AAT_I superfamily) |
|||
|
PHATRDRAFT_41003 : (COG5134 superfamily) |
PHATRDRAFT_49058 : metal transport protein |
||
|
PHATRDRAFT_6789 : peroxisomal coenzyme a diphosphatase nudt7 (nucleoside diphosphate-linked moiety x motif 7) (nudix motif 7) (CoAse) |
PHATRDRAFT_9200 : serine threonine kinase 4 (PKc_like superfamily) |
PHATRDRAFT_16827 : unusual protein kinase (AarF) |
PHATRDRAFT_33394 : membrane protein (SYS1 superfamily) |
|
PHATRDRAFT_47991 : asc-1 complex subunit p100 |
PHATRDRAFT_33380 : heat shock factor 3 (HSF_DNA-bind) |
PHATRDRAFT_16105 : transmembrane proteinisoform cra_a (Solute_trans_a) |
PHATRDRAFT_36359 : undecaprenyl diphosphate synthase (Cis_IPPS) |
|
PHATRDRAFT_5288 : (AANH_like superfamily) |
PHATRDRAFT_35200 : nadh dehydrogenase1 alphaassembly factor 1 (CIA30 superfamily) |
PHATRDRAFT_46645 : transmembrane protein 34 (Solute_trans_a superfamily) |
PHATRDRAFT_11352 : membrance occupation and recognition nexus protein 1 (MORN superfamily) |
|
PHATRDRAFT_44797 : taf9 rna polymerase ii tata box binding protein-associated factor (TAF9) |
PHATRDRAFT_15870 : dreg-2 like protein (DREG-2 superfamily) |
PHATRDRAFT_48117 : ntp7b_xenlasodium bile acid cotransporter 7-b (na(+) bile acid cotransporter 7-b) (solute carrier family 10 member 7-b) (DUF4137) |
|
|
PHATRDRAFT_5099 : necap endocytosis associated 2 (PHear_NECAP) |
PHATRDRAFT_46865 : (AlkA) |
PHATRDRAFT_44799 : had-superfamily hydrolase subfamilyvariant 1 and 3 (COG1011) |
PHATRDRAFT_42484 : golgi traffic protein (Got1) |
|
PHATRDRAFT_14003 : protein kinase (S_TKc) |
PHATRDRAFT_50605 : capsular polysaccharide biosynthesisprotein (DUF563) |
PHATRDRAFT_52661 : pci domain containing protein |
|
|
PHATRDRAFT_51511 : member ras oncogene family (Rab5_related) |
PHATRDRAFT_3296 : glycine d-amino acid oxidases-like |
PHATRDRAFT_4634 : dhhc zinc finger domain containing protein (zf-DHHC) |
|
|
PHATRDRAFT_45874 : zinc finger (c3hc4-type ring finger) family protein (RING) |
PHATRDRAFT_45606 : mitochondrial inner membrane metallopeptidase (Peptidase_M48) |
PHATRDRAFT_37632 : conserved membrane protein (VIT1) |
|
|
PHATRDRAFT_19669 : vacuolar protein sorting 28 (VPS28 superfamily) |
PHATRDRAFT_40152 : (SAYSvFN superfamily) |
PHATRDRAFT_52208 : alcohol dehydrogenasedomain protein (Zn_ADH5) |
|
|
PHATRDRAFT_5508 : kin17 cg5649-pa (Kin17_mid) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.480 | 0.000505051 |
| Copper_SH | Copper_SH | -0.323 | 0.000515464 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.018 | 0.870595 |
| Silver_SH | Silver_SH | -0.183 | 0.00637584 |
| Cadmium_1.2mg | Cadmium_1.2mg | -0.015 | 0.824808 |
| BlueLight_24h | BlueLight_24h | 0.061 | 0.843814 |
| GreenLight_0.5h | GreenLight_0.5h | 1.151 | 0.000458716 |
| RedLight_6h | RedLight_6h | 0.279 | 0.0508427 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.980594 |
| GreenLight_24h | GreenLight_24h | 0.138 | 0.21612 |
| light_6hr | light_6hr | 0.289 | 0.0356771 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.213 | 0.0852657 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.030 | 0.70739 |
| Ammonia_SH | Ammonia_SH | 0.499 | 0.00350877 |
| Simazine_SH | Simazine_SH | -0.107 | 0.180563 |
| Oil_SH | Oil_SH | -0.053 | 0.422294 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.103 | 0.188 |
| Cadmium_SH | Cadmium_SH | 0.206 | 0.00992908 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.096 | 0.366531 |
| highlight_0to6h | highlight_0to6h | 0.149 | 0.14491 |
| BlueLight_0.5h | BlueLight_0.5h | 1.609 | 0.000462963 |
| Dispersant_SH | Dispersant_SH | 0.003 | 0.975915 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.134 | 0.0779255 |
| light_15.5hr | light_15.5hr | -0.110 | 0.526284 |
| light_10.5hr | light_10.5hr | 0.240 | 0.14089 |
| Dispersed_oil_SH | Dispersed_oil_SH | 0.011 | 0.892708 |
| Si_free | Si_free | 0.118 | 0.274699 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 1.657 | 0.000396825 |
| Mixture_SH | Mixture_SH | 0.415 | 0.0203863 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.147 | 0.261859 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.209 | 0.16359 |
| BlueLight_6h | BlueLight_6h | 0.426 | 0.000581395 |
| highlight_12to48h | highlight_12to48h | -0.138 | 0.341304 |
| RedLight_24h | RedLight_24h | 0.157 | 0.140206 |
| Dark_treated | Dark_treated | 1.608 | 0.000446429 |
| GreenLight_6h | GreenLight_6h | 0.456 | 0.000609756 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.023 | 0.4 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.077 | 0.106373 |
| RedLight_0.5h | RedLight_0.5h | 0.837 | 0.000555556 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.177 | 0.14633 |
| Re-illuminated_24h | Re-illuminated_24h | 0.243 | 0.00119048 |