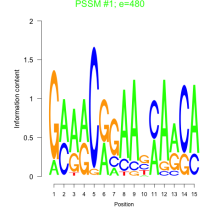
Phatr_bicluster_0117 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0117 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12234 nucleotidyltransferase family protein (PAP_central)
PHATRDRAFT_13294 ribosomal large subunit pseudouridine synthase a (RluA)
PHATRDRAFT_16065 shwachman-bodian-diamond syndrome (COG1500)
PHATRDRAFT_16155 yl92_schpo uncharacterized protein (PTZ00368)
PHATRDRAFT_16560 3-5 exoribonuclease ph component of the exosome (RNase_PH superfamily)
PHATRDRAFT_18274 cyclophilin-like protein (cyclophilin_ABH_like)
PHATRDRAFT_19659 alcoholiron1 isoform 1 (HOT)
PHATRDRAFT_29958 u2 snrnp auxiliarysmall subunit (RRM_SF superfamily)
PHATRDRAFT_3182 hect (homologous to the e6-apcarboxyl terminus) domain and rcc1-like domainpartial (RCC1)
PHATRDRAFT_33524 cax-interacting protein 4-like
PHATRDRAFT_37109 transmembrane efflux protein (ATG22)
PHATRDRAFT_40731 amino acid permease-like protein (Aa_trans superfamily)
PHATRDRAFT_45173 tpa: zinc finger protein (zf-DHHC)
PHATRDRAFT_45199 serine arginine repetitive matrix 2 (PTZ00121)
PHATRDRAFT_45412 (ZnMc superfamily)
PHATRDRAFT_45968 nucleosome binding protein (HMGB-UBF_HMG-box)
PHATRDRAFT_48350 (CHD)
PHATRDRAFT_5060 transcription factor dp-2 (e2f dimerization partner 2) (DP superfamily)
PHATRDRAFT_6032 diguanylate cyclase with gaf sensor (GAF)
PHATRDRAFT_6387 membrane family (SapB)
PHATRDRAFT_7007 ntp pyrophosphohydrolase (Nudix_Hydrolase superfamily)
PHATRDRAFT_7026 ck1 family protein kinase (PKc_like superfamily)
PHATRDRAFT_8706 6-phosphofructo-2-kinase fructose--bisphosphatase short form (AAA_17 superfamily)
PHATRDRAFT_8737 98b protein (HMGB-UBF_HMG-box)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006003 | fructose 2,6-bisphosphate metabolism | 0.00887241 | 1 | Phatr_bicluster_0117 |
| GO:0006396 | RNA processing | 0.0249465 | 1 | Phatr_bicluster_0117 |
| GO:0006807 | nitrogen compound metabolism | 0.0605653 | 1 | Phatr_bicluster_0117 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0698674 | 1 | Phatr_bicluster_0117 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0730847 | 1 | Phatr_bicluster_0117 |
| GO:0006350 | transcription | 0.0895323 | 1 | Phatr_bicluster_0117 |
| GO:0006865 | amino acid transport | 0.117654 | 1 | Phatr_bicluster_0117 |
| GO:0000074 | regulation of cell cycle | 0.141104 | 1 | Phatr_bicluster_0117 |
| GO:0007242 | intracellular signaling cascade | 0.171456 | 1 | Phatr_bicluster_0117 |
| GO:0007165 | signal transduction | 0.189908 | 1 | Phatr_bicluster_0117 |


Comments